6KS6
 
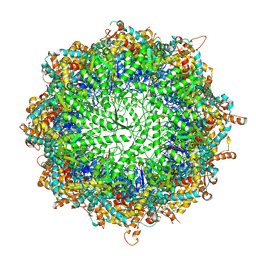 | | TRiC at 0.2 mM ADP-AlFx, Conformation 1, 0.2-C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KVH
 
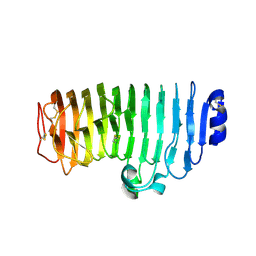 | | The mutant crystal structure of endo-polygalacturonase (T284A) from Talaromyces leycettanus JCM 12802 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose, endo-polygalacturonase | | Authors: | Tu, T, Wang, Z, Luo, H, Yao, B. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Insights into the Mechanisms Underlying the Kinetic Stability of GH28 Endo-Polygalacturonase.
J.Agric.Food Chem., 69, 2021
|
|
7E7X
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
4Z8M
 
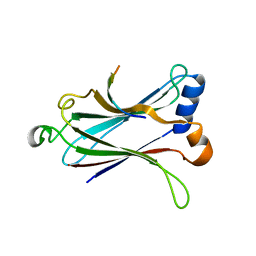 | | Crystal structure of the MAVS-TRAF6 complex | | Descriptor: | Peptide from Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 6 | | Authors: | Shi, Z.B, Zhou, Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Mitochondrial Antiviral Signaling Protein (MAVS)-Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) Signaling
J.Biol.Chem., 290, 2015
|
|
6L31
 
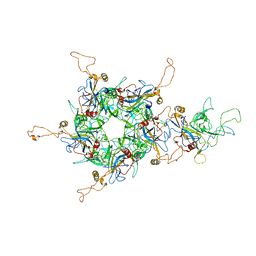 | | L1 protein of human papillomavirus 6 | | Descriptor: | Major capsid protein L1 | | Authors: | Li, S.W, Liu, X.L, Gu, Y. | | Deposit date: | 2019-10-07 | | Release date: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Neutralization sites of human papillomavirus-6 relate to virus attachment and entry phase in viral infection.
Emerg Microbes Infect, 8, 2019
|
|
7E39
 
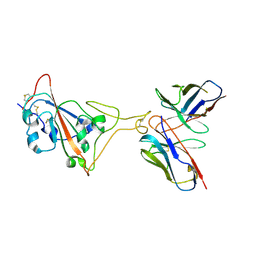 | | SARS-CoV-2 spike in complex with the Ab4 neutralizing antibody (State 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Ab4, Light Chain of Ab4, ... | | Authors: | Liu, C. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Three epitope-distinct human antibodies from RenMab mice neutralize SARS-CoV-2 and cooperatively minimize the escape of mutants.
Cell Discov, 7, 2021
|
|
7E3B
 
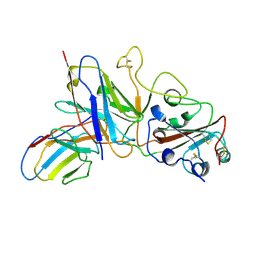 | |
7E3C
 
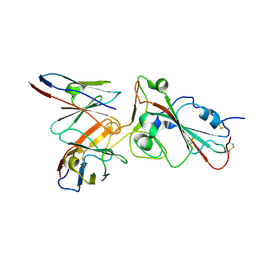 | |
8QEO
 
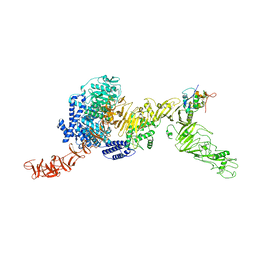 | |
8QEN
 
 | |
7E5R
 
 | | SARS-CoV-2 S trimer with three-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
8U1F
 
 | | FGFR2 Kinase Domain Bound to Irreversible Inhibitor Cmpd 10 | | Descriptor: | Fibroblast growth factor receptor 2, GLYCEROL, N-[4-(4-amino-7-methyl-5-{4-[(4-methylpyrimidin-2-yl)oxy]phenyl}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)phenyl]-2-methylpropanamide, ... | | Authors: | Valverde, R, Foster, L. | | Deposit date: | 2023-08-31 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7E5S
 
 | | SARS-CoV-2 S trimer with four-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
5C6D
 
 | | Crystal structure of USP7 in complex with UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Zhang, Z.-M, Song, J. | | Deposit date: | 2015-06-22 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1.
Cell Rep, 12, 2015
|
|
8JBJ
 
 | | Crystal structure of anti-PVRIG Fab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Li, X.L, Song, J. | | Deposit date: | 2023-05-09 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification and characterization of an unexpected isomerization motif in CDRH2 that affects antibody activity.
Mabs, 15, 2023
|
|
6MPH
 
 | | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DF1W-a.01 Light chain, ... | | Authors: | Acharya, P, Xu, K, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
4MZ9
 
 | | Revised structure of E. coli SSB | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Oakley, A.J. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy.
Nucleic Acids Res., 42, 2014
|
|
6MX8
 
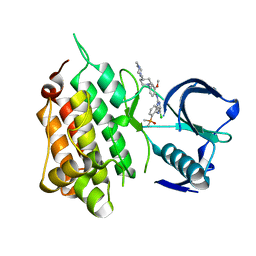 | | Crystal structure of anaplastic lymphoma kinase (ALK) bound by Brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, ALK tyrosine kinase receptor | | Authors: | Dougan, D.R, Zhou, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Brigatinib (AP26113), a Phosphine Oxide-Containing, Potent, Orally Active Inhibitor of Anaplastic Lymphoma Kinase.
J. Med. Chem., 59, 2016
|
|
6KDV
 
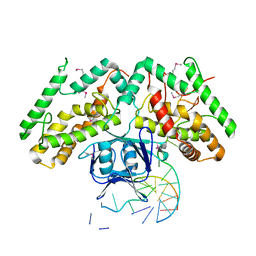 | | Crystal structure of TtCas1-DNA complex | | Descriptor: | CRISPR-associated endonuclease Cas1 2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*GP*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*CP*CP*AP*GP*CP*AP*TP*CP*GP*AP*CP*TP*C)-3') | | Authors: | Wang, Y.L, Li, J.Z, Yang, J, Wang, J.Y. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-18 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of Cas1 in complex with branched DNA.
Sci China Life Sci, 63, 2020
|
|
5CH8
 
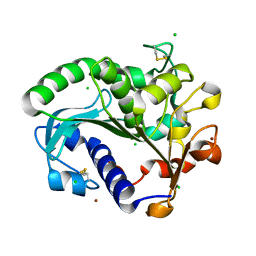 | | Crystal structure of MDLA N225Q mutant form Penicillium cyclopium | | Descriptor: | CHLORIDE ION, GLYCEROL, Mono- and diacylglycerol lipase, ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-07-10 | | Release date: | 2016-04-20 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Lipase-Driven Epoxidation Is A Two-Stage Synergistic Process
ChemistrySelect, 4, 2016
|
|
