8H59
 
 | | A fungal MAP kinase in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase MPS1, ~{N}-[(2~{S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]-8-[2-methoxy-5-(trifluoromethyloxy)phenyl]-1,6-naphthyridine-2-carboxamide | | Authors: | Kong, Z, Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Aided Identification of an Inhibitor Targets Mps1 for the Management of Plant-Pathogenic Fungi.
Mbio, 14, 2023
|
|
7E7G
 
 | |
7EXM
 
 | | The N-terminal crystal structure of SARS-CoV-2 NSP2 | | Descriptor: | GLYCEROL, Non-structural protein 2, ZINC ION | | Authors: | Ma, J, Chen, Z. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure and Function of N-Terminal Zinc Finger Domain of SARS-CoV-2 NSP2.
Virol Sin, 36, 2021
|
|
8ILH
 
 | | The crystal structure of dG(Se-Sp)-DNA:Pol X product binary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOSELENOPHOSPHATE, DNA (5'-D(*CP*GP*GP*AP*TP*CP*C)-3'), MAGNESIUM ION, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILI
 
 | | The crystal structure of dG(Se-Rp)-DNA:Pol X product binary complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*TP*CP*CP*(7S8))-3'), MAGNESIUM ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILE
 
 | | The crystal structure of dGTPalphaSe-Rp:DNApre-II:Pol X substrate ternary complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), MANGANESE (II) ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILG
 
 | | The crystal structure of dG-DNA:Pol X product binary complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*TP*CP*CP*G)-3'), FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILF
 
 | | The crystal structure of dGTPalphaSe-Sp:DNApre-II:Pol X substrate ternary complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), MANGANESE (II) ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Zhao, Q.W, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
7XC5
 
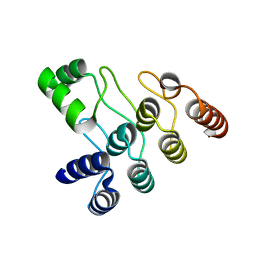 | | Crystal structure of the ANK domain of CLPB | | Descriptor: | Isoform 2 of Caseinolytic peptidase B protein homolog | | Authors: | Liu, Y, Wu, D, Lu, G, Gao, N, Lin, J. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
7VNQ
 
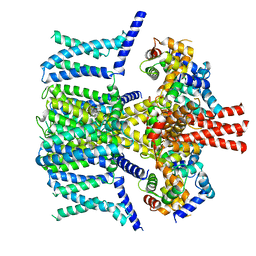 | | Structure of human KCNQ4-ML213 complex in nanodisc | | Descriptor: | (1S,2S,4R)-N-(2,4,6-trimethylphenyl)bicyclo[2.2.1]heptane-2-carboxamid, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Xu, F, Zheng, Y. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into the lipid and ligand regulation of a human neuronal KCNQ channel.
Neuron, 110, 2022
|
|
7VNR
 
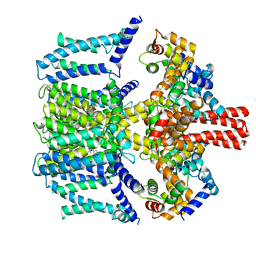 | | Structure of human KCNQ4-ML213 complex in digitonin | | Descriptor: | (1S,2S,4R)-N-(2,4,6-trimethylphenyl)bicyclo[2.2.1]heptane-2-carboxamid, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Xu, F, Zheng, Y. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the lipid and ligand regulation of a human neuronal KCNQ channel.
Neuron, 110, 2022
|
|
7VNP
 
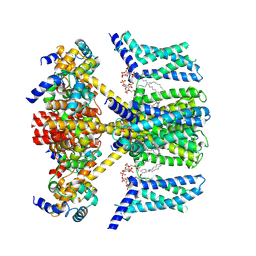 | | Structure of human KCNQ4-ML213 complex with PIP2 | | Descriptor: | (1S,2S,4R)-N-(2,4,6-trimethylphenyl)bicyclo[2.2.1]heptane-2-carboxamid, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Xu, F, Zheng, Y. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural insights into the lipid and ligand regulation of a human neuronal KCNQ channel.
Neuron, 110, 2022
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7BZH
 
 | |
7CQQ
 
 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQL
 
 | | Apo GmaS without ligand | | Descriptor: | Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CM9
 
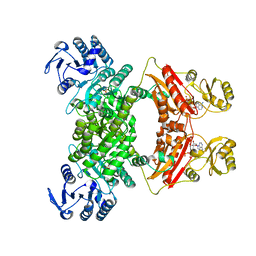 | | DMSP lyase DddX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DMSP lyase, SULFATE ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-07-25 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.
Elife, 10, 2021
|
|
7F38
 
 | |
7XC2
 
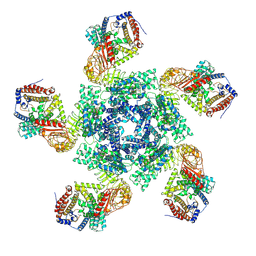 | | Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Avirulence factor, CNL9 | | Authors: | Alexander, F, Li, E.T, Aaron, L, Deng, Y.N, Sun, Y, Chai, J.J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A wheat resistosome defines common principles of immune receptor channels.
Nature, 610, 2022
|
|
7CQX
 
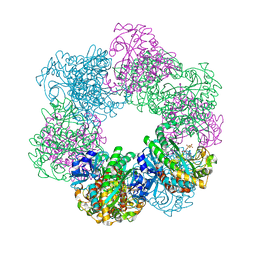 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQW
 
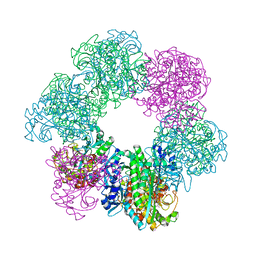 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7D4N
 
 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 20 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
7D4M
 
 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 5 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
