6DFP
 
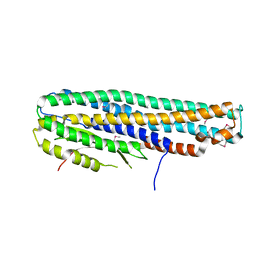 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | Descriptor: | VCA0883 | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
4W9Y
 
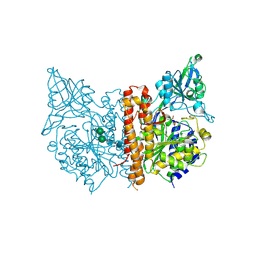 | |
6O3Z
 
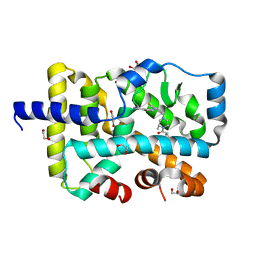 | | Crystal structure of RORgt with 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide, RAR-related orphan receptor C isoform a variant | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of [1,2,4]Triazolo[1,5-a]pyridine Derivatives as Potent and Orally Bioavailable ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 11, 2020
|
|
5BQD
 
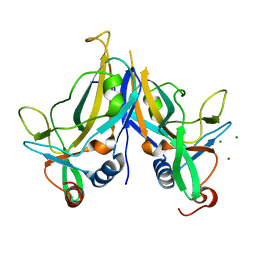 | | Crystal Structure of TBX5 (1-239) Dimer | | Descriptor: | MAGNESIUM ION, T-box transcription factor TBX5 | | Authors: | Pradhan, L, Gopal, S, Patel, A, Kasahara, H, Nam, H.J. | | Deposit date: | 2015-05-28 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5.
Biochemistry, 55, 2016
|
|
6RKJ
 
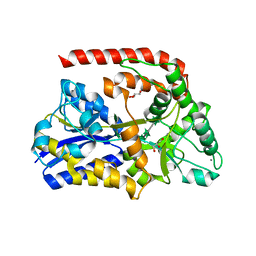 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinooctaose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RKH
 
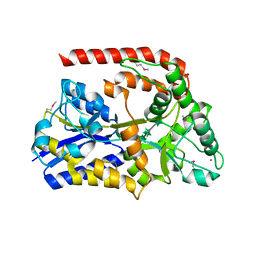 | | The crystal structure of AbnE (Selenium derivative), an arabino-oligosaccharide binding protein, in complex with arabinohexaose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RL2
 
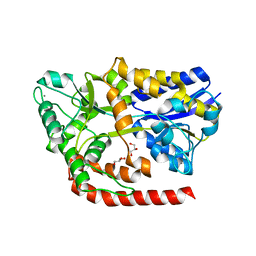 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinotetraose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6IWB
 
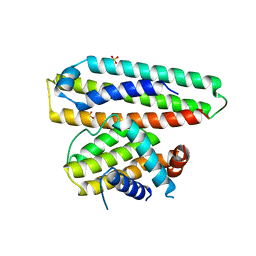 | | Crystal structure of a computationally designed protein (LD3) in complex with BCL-2 | | Descriptor: | Apolipoprotein E, Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, SULFATE ION | | Authors: | Kim, S, Kwak, M.J, Oh, B.-H, Correia, B.E, Gainza, P. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy.
Nat.Biotechnol., 38, 2020
|
|
6RJY
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arabino-oligosaccharids-binding protein, CALCIUM ION, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RKL
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinoheptaose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RL1
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinotriose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RKX
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinopentaose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6O3T
 
 | | Structural basis of FOXC2 and DNA interactions | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein C2 | | Authors: | Nam, H.-J, Li, S. | | Deposit date: | 2019-02-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal Structure of FOXC2 in Complex with DNA Target.
Acs Omega, 4, 2019
|
|
6K0T
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-17 | | Descriptor: | 3-[(1~{E})-1-[8-[(8-chloranyl-2-cyclopropyl-imidazo[1,2-a]pyridin-3-yl)methyl]-3-fluoranyl-6~{H}-benzo[c][1]benzoxepin-11-ylidene]ethyl]-4~{H}-1,2,4-oxadiazol-5-one, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Suzuki, M, Yamamoto, K, Takahashi, Y, Saito, J. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-30 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Development of a novel class of peroxisome proliferator-activated receptor (PPAR) gamma ligands as an anticancer agent with a unique binding mode based on a non-thiazolidinedione scaffold.
Bioorg.Med.Chem., 27, 2019
|
|
6KQQ
 
 | | NSD1 SET domain in complex with BT3 and SAM | | Descriptor: | 2-azanyl-6-[(2-azanyl-4-oxidanyl-1,3-benzothiazol-6-yl)disulfanyl]-1,3-benzothiazol-4-ol, 2-azanyl-6-sulfanyl-1,3-benzothiazol-4-ol, CALCIUM ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2019-08-18 | | Release date: | 2020-09-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent inhibition of NSD1 histone methyltransferase.
Nat.Chem.Biol., 16, 2020
|
|
6RTI
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with aptamer A9g | | Descriptor: | (2S)-2-(PHOSPHONOMETHYL)PENTANEDIOIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Kolenko, P, Barinka, C. | | Deposit date: | 2019-05-24 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of prostate-specific membrane antigen recognition by the A9g RNA aptamer.
Nucleic Acids Res., 48, 2020
|
|
6KQP
 
 | | NSD1 SET domain in complex with SAM | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2019-08-18 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Covalent inhibition of NSD1 histone methyltransferase.
Nat.Chem.Biol., 16, 2020
|
|
5X79
 
 | | Human GST Pi conjugated with novel inhibitor, GS-ESF | | Descriptor: | (2S)-2-azanyl-5-[[(2R)-3-(2-fluorosulfonylethylsulfanyl)-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase P | | Authors: | Tomoike, F, Shishido, Y, Fukui, K, Kimura, Y, Abe, H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A covalent G-site inhibitor for glutathione S-transferase Pi (GSTP1-1).
Chem. Commun. (Camb.), 53, 2017
|
|
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | Descriptor: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
4XQ0
 
 | |
4XPZ
 
 | |
5JUE
 
 | | Crystal Structure of UIC2 Fab | | Descriptor: | GLYCEROL, heavy chain of UIC2 Fab, light chain of UIC2 Fab | | Authors: | Xia, D, Esser, L. | | Deposit date: | 2016-05-10 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the antigen-binding fragment of a monoclonal antibody specific for the multidrug-resistance-linked ABC transporter human P-glycoprotein.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6W08
 
 | | Crystal Structure of Motility Associated Killing Factor E from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Joachimiak, G, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | Authors: | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-25 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
5KO2
 
 | | Mouse pgp 34 linker deleted mutant Hg derivative | | Descriptor: | MERCURY (II) ION, Multidrug resistance protein 1A | | Authors: | Xia, D, Esser, L, Zhou, F. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity.
J. Biol. Chem., 292, 2017
|
|
