6AWM
 
 | | PawL-Derived Peptide PLP-4 | | Descriptor: | GLY-LEU-LEU-GLY-ILE-THR-ASP | | Authors: | Fisher, M, Mylne, J.S, Howard, M.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A family of small, cyclic peptides buried in preproalbumin since the Eocene epoch.
Plant Direct, 2, 2018
|
|
3SRQ
 
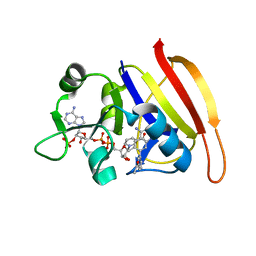 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 1-[3-(2,4-diamino-6-methylquinazolin-7-yl)phenyl]ethanone, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5WA5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor XMD11-50 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2017-06-24 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
2K3N
 
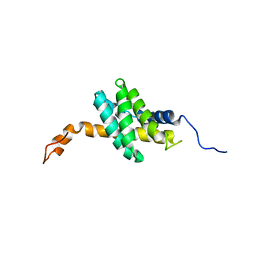 | |
2K3O
 
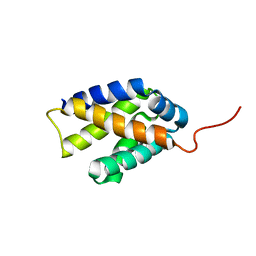 | |
3TUO
 
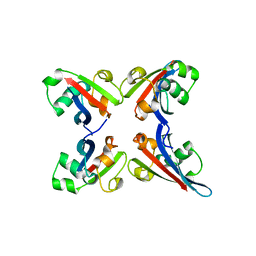 | |
2K3Q
 
 | |
5ZFW
 
 | | Crystal structure of human DUX4 homeodomains bound to A11G DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*GP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*CP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
3RER
 
 | | Crystal structure of E. coli Hfq in complex with AU6A RNA and ADP | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*UP*A)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, W.W, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2011-04-05 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cooperation of Escherichia coli Hfq hexamers in DsrA binding.
Genes Dev., 25, 2011
|
|
5ZFY
 
 | | Crystal structure of human DUX4 homeodomains bound to A12C DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*CP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*GP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
2K3P
 
 | |
4ONM
 
 | | Crystal structure of human Mms2/Ubc13 - NSC697923 | | Descriptor: | 2-[(4-methylphenyl)sulfonyl]-5-nitrofuran, GLYCEROL, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4OUM
 
 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CITRATE ANION, Caprin-2, ISOPROPYL ALCOHOL, ... | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
3CWD
 
 | | Molecular recognition of nitro-fatty acids by PPAR gamma | | Descriptor: | (9E,12Z)-10-nitrooctadeca-9,12-dienoic acid, (9Z,12E)-12-nitrooctadeca-9,12-dienoic acid, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Martynowski, D, Li, Y. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of nitrated fatty acids by PPAR gamma.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5DT2
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD11 [N4-methyl-N2-(2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl)pyrimidine-2,4-diamine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~4~-methyl-N~2~-[2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl]pyrimidine-2,4-diamine, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DRY
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [N-(1-(2-chlorophenyl)-1H-indol-6-yl)-2-(2-(5-(2-chlorophenyl)-1H-tetrazol-1-yl)acetyl)hydrazinecarboxamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-[1-(2-chlorophenyl)-1H-indol-6-yl]-2-{[5-(2-chlorophenyl)-1H-tetrazol-1-yl]acetyl}hydrazinecarboxamide, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5H9T
 
 | | Crystal structure of native NalD at resolution of 2.9, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | Descriptor: | NalD | | Authors: | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa.
Mol. Microbiol., 100, 2016
|
|
2MBK
 
 | | The Clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed | | Descriptor: | Crossveinless 2 | | Authors: | Mueller, T.D, Fiebig, J.E, Weidauer, S.E, Qiu, L, Bauer, M, Schmieder, P, Beerbaum, M, Zhang, J, Oschkinat, H, Sebald, W. | | Deposit date: | 2013-08-02 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Clip-Segment of the von Willebrand Domain 1 of the BMP Modulator Protein Crossveinless 2 Is Preformed.
Molecules, 18, 2013
|
|
4L2Z
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
5EEG
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with tetrazole-SAH | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[[(3~{S})-3-azanyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)propyl]sulfanylmethyl]oxolane-3,4-diol, Carminomycin 4-O-methyltransferase DnrK | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
5HMM
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exodeoxyribonuclease, ... | | Authors: | Flemming, C.S, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8J0K
 
 | | Crystal structure of human TFAP2A in complex with DNA | | Descriptor: | DNA (5'-D(*CP*TP*GP*CP*CP*TP*CP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*CP*GP*AP*GP*GP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | Deposit date: | 2023-04-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
8J0Q
 
 | | Structure of DNA binding domain of human TFAP2B | | Descriptor: | GLYCEROL, Transcription factor AP-2-beta | | Authors: | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | Deposit date: | 2023-04-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
8J0L
 
 | | Structure of DNA binding Domain of Human TFAP2A | | Descriptor: | GLYCEROL, Transcription factor AP-2-alpha | | Authors: | Liu, K, Xiao, Y.Q, Gan, L.Y, Min, J.R. | | Deposit date: | 2023-04-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
7CTT
 
 | | Cryo-EM structure of Favipiravir bound to replicating polymerase complex of SARS-CoV-2 in the pre-catalytic state. | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Peng, Q, Peng, R, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of SARS-CoV-2 Polymerase Inhibition by Favipiravir.
Innovation (N Y), 2, 2021
|
|
