3NII
 
 | | The structure of UBR box (KIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIM
 
 | | The structure of UBR box (RRAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RRAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIJ
 
 | | The structure of UBR box (HIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIN
 
 | | The structure of UBR box (RLGES) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RLGES, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIT
 
 | | The structure of UBR box (native1) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
6G3C
 
 | | Crystal Structure of JAK2-V617F pseudokinase domain in complex with Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]amino]-5,7,7-trimethyl-8-(3-methylbutyl)pteridin-6-one, Tyrosine-protein kinase | | Authors: | Dekker, C, Hinniger, A. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
Acs Chem.Biol., 14, 2019
|
|
3NIH
 
 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIL
 
 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
8FV3
 
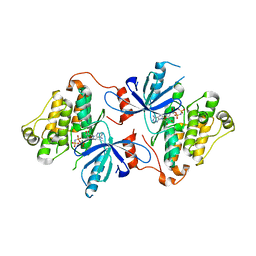 | | EGFR(T790M/V948R) in complex with compound 1 (LN4503) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(4P)-4-[(4P)-5-{3-[(8-fluoro-11-oxo-5,11-dihydro-10H-dibenzo[b,e][1,4]diazepin-10-yl)methyl]phenyl}-2-(methylsulfanyl)-1H-imidazol-4-yl]pyridin-2-yl}acetamide, ... | | Authors: | Ogboo, B.C, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
1HCT
 
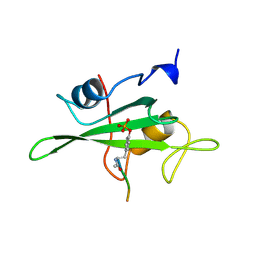 | |
1HCS
 
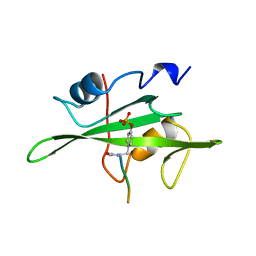 | |
2TNF
 
 | | 1.4 A RESOLUTION STRUCTURE OF MOUSE TUMOR NECROSIS FACTOR, TOWARDS MODULATION OF ITS SELECTIVITY AND TRIMERISATION | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, PROTEIN (TUMOR NECROSIS FACTOR ALPHA) | | Authors: | Baeyens, K.J, De Bondt, H.L, Raeymaekers, A, Fiers, W, De Ranter, C.J. | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of mouse tumour-necrosis factor at 1.4 A resolution: towards modulation of its selectivity and trimerization.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7TXH
 
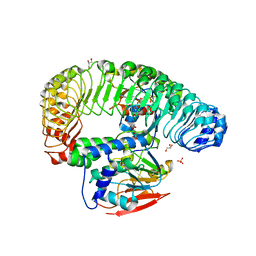 | | Human MRas Q71R in complex with human Shoc2 LRR domain M173I and human PP1Ca | | Descriptor: | GLYCEROL, Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, ... | | Authors: | Hauseman, Z.J, Viscomi, J, Dhembi, A, Clark, K, King, D.A, Fodor, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the MRAS-SHOC2-PP1C phosphatase complex.
Nature, 609, 2022
|
|
7TYG
 
 | |
7UKW
 
 | | EGFR(T790M/V948R) in complex with Lazertinib (YH25448) | | Descriptor: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | Authors: | Pham, C.D, Heppner, D.E. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
4F9G
 
 | | Crystal structure of STING complex with Cyclic di-GMP. | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Transmembrane protein 173 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cyclic di-GMP Sensing via the Innate Immune Signaling Protein STING.
Mol.Cell, 46, 2012
|
|
4F9E
 
 | |
2HWO
 
 | |
2HWP
 
 | |
1QQG
 
 | |
5TWL
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 9-(3,5-dichloro-4-hydroxyphenyl)-1-{trans-4-[(dimethylamino)methyl]cyclohexyl}-3,4-dihydropyrimido[5,4-c]quinolin-2(1H)-one, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWZ
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-{[2-methoxy-4-(1H-pyrazol-4-yl)benzoyl]amino}-2,3,4,5-tetrahydro-1H-3-benzazepinium, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWY
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 2-(benzyloxy)-4-(1H-pyrazol-4-yl)-N-(2,3,4,5-tetrahydro-1H-3-benzazepin-7-yl)benzamide, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
4LGH
 
 | |
4LGG
 
 | | Structure of 3MB-PP1 bound to analog-sensitive Src kinase | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lopez, M.S, Dar, A.C, Shokat, K.M. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology.
Acs Chem.Biol., 8, 2013
|
|
