1ASQ
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
4WX7
 
 | | Crystal structure of adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | 3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-N-(2-{[(2Z)-2-iminoethyl]amino}-2-oxoethyl)-4-methoxybenzamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1ASP
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
5P21
 
 | |
11BG
 
 | | A POTENTIAL ALLOSTERIC SUBSITE GENERATED BY DOMAIN SWAPPING IN BOVINE SEMINAL RIBONUCLEASE | | Descriptor: | PROTEIN (BOVINE SEMINAL RIBONUCLEASE), SULFATE ION, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Adinolfi, S, Sica, F, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 1999-03-11 | | Release date: | 1999-11-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potential allosteric subsite generated by domain swapping in bovine seminal ribonuclease.
J.Mol.Biol., 293, 1999
|
|
8OS5
 
 | | Crystal structure of the Factor XII heavy chain reveals an interlocking dimer with a FnII to kringle domain interaction | | Descriptor: | Coagulation factor XII-Mie | | Authors: | Li, C, Saleem, M, Kaira, B.G, Brown, A, Wilson, C, Philippou, H, Emsley, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Factor XII and kininogen asymmetric assembly with gC1qR/C1QBP/P32 is governed by allostery.
Blood, 136, 2020
|
|
8P6U
 
 | | Human carbonic anhydrase II containing 5-fluorotryptophanes | | Descriptor: | BENZOIC ACID, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
8PHL
 
 | | Human carbonic anhydrase II containing 4-fluorophenylalanine | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-06-20 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
1AOZ
 
 | | REFINED CRYSTAL STRUCTURE OF ASCORBATE OXIDASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Ladenstein, R, Huber, R. | | Deposit date: | 1992-01-08 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined crystal structure of ascorbate oxidase at 1.9 A resolution.
J.Mol.Biol., 224, 1992
|
|
1AIF
 
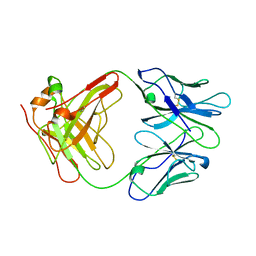 | | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB FROM MOUSE | | Descriptor: | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB (HEAVY CHAIN), ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB (LIGHT CHAIN) | | Authors: | Ban, N, Escobar, C, Hasel, K, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-11-14 | | Release date: | 1997-02-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an anti-idiotypic Fab against feline peritonitis virus-neutralizing antibody and a comparison with the complexed Fab.
FASEB J., 9, 1995
|
|
8I2Z
 
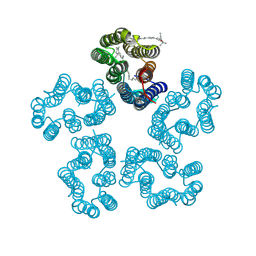 | | Cryo-EM structure of the zeaxanthin-bound kin4B8 | | Descriptor: | RETINAL, Xanthorhodopsin, Zeaxanthin | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2023-01-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
8OHZ
 
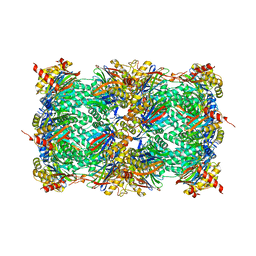 | | Yeast 20S proteasome in complex with a photoswitchable cepafungin derivative (transCep1) | | Descriptor: | (2~{S},3~{R})-2-[2-[4-[2-(4-ethylphenyl)hydrazinyl]phenyl]ethanoylamino]-~{N}-[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]-3-oxidanyl-butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Morstein, J, Amatuni, A, Schuster, A, Kuttenlochner, W, Ko, T, Groll, M, Adibekian, A, Renata, H, Trauner, D.H. | | Deposit date: | 2023-03-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OI1
 
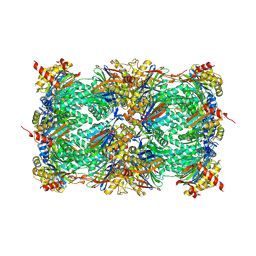 | | Yeast 20S proteasome in complex with a photoswitchable cepafungin derivative (transCep4) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Morstein, J, Amatuni, A, Schuster, A, Kuttenlochner, W, Ko, T, Groll, M, Adibekian, A, Renata, H, Trauner, D.H. | | Deposit date: | 2023-03-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OO2
 
 | | ChdA complex with amido-chelocardin | | Descriptor: | 2-carboxamido-2-deacetyl-chelocardin, MAGNESIUM ION, Putative transcriptional regulator | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Revision of the Absolute Configurations of Chelocardin and Amidochelocardin.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2JOK
 
 | | NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2JNP
 
 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-3, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JQM
 
 | | Yellow Fever Envelope Protein Domain III NMR Structure (S288-K398) | | Descriptor: | Envelope protein E | | Authors: | Volk, D.E, Gandham, S.H, May, F.J, Anderson, A, Barrett, A.D, Gorenstein, D.G. | | Deposit date: | 2007-06-03 | | Release date: | 2008-06-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of yellow fever virus envelope protein domain III.
Virology, 394, 2009
|
|
2JC0
 
 | | CRYSTAL STRUCTURE OF HEPATITIS C VIRUS POLYMERASE IN COMPLEX WITH INHIBITOR SB655264 | | Descriptor: | (2S,4S,5R)-2-ISOBUTYL-5-(2-THIENYL)-1-[4-(TRIFLUOROMETHYL)BENZOYL]PYRROLIDINE-2,4-DICARBOXYLIC ACID, RNA-DEPENDENT RNA-POLYMERASE | | Authors: | Wonacott, A, Skarzynski, T, Singh, O.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of Novel Acyl Pyrrolidine Inhibitors of Hepatitis C Virus RNA-Dependent RNA Polymerase Leading to a Development Candidate.
J.Med.Chem., 50, 2007
|
|
2JFT
 
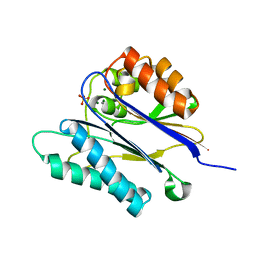 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis in complex with sulfate | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, SER-THR PHOSPHATASE MSPP, ... | | Authors: | Bellinzoni, M, Wehenkel, A, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Insights Into the Mechanism of Ppm Ser/Thr Phosphatases from the Atomic Resolution Structures of a Mycobacterial Enzyme
Structure, 15, 2007
|
|
2JFR
 
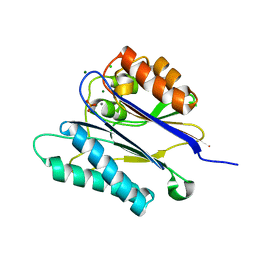 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis in complex with phosphate at 0.83 A resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bellinzoni, M, Wehenkel, A, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Insights Into the Mechanism of Ppm Ser/Thr Phosphatases from the Atomic Resolution Structures of a Mycobacterial Enzyme
Structure, 15, 2007
|
|
2JHN
 
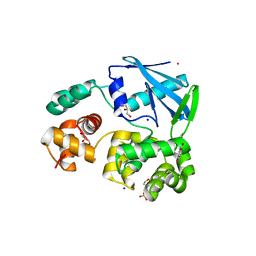 | | 3-methyladenine dna-glycosylase from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-METHYLADENINE DNA-GLYCOSYLASE, GLYCEROL, ... | | Authors: | Leiros, I, Nabong, M.P, Grosvik, K, Ringvoll, J, Haugland, G.T, Uldal, L, Reite, K, Olsbu, I.K, Knaevelsrud, I, Moe, E, Andersen, O.A, Birkeland, N.K, Ruoff, P, Klungland, A, Bjelland, S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Enzymatic Excision of N1-Methyladenine and N3-Methylcytosine from DNA
Embo J., 26, 2007
|
|
2JJK
 
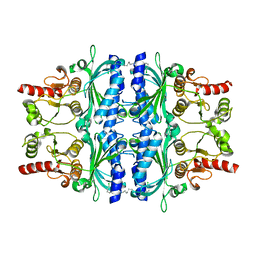 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE -1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH A DUAL BINDING AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N,N'-(heptane-1,7-diyldicarbamoyl)bis(3-chlorobenzenesulfonamide) | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Hebeisen, P. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Fbpase Inhibitors Gain 10(5) Times in Potency When Simultaneously Binding Two Neighboring AMP Sites.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2JC1
 
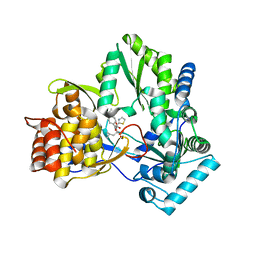 | | CRYSTAL STRUCTURE OF HEPATITIS C VIRUS POLYMERASE IN COMPLEX WITH INHIBITOR SB698223 | | Descriptor: | (2S,4S,5R)-1-(4-TERT-BUTYLBENZOYL)-2-ISOBUTYL-5-(1,3-THIAZOL-2-YL)PYRROLIDINE-2,4-DICARBOXYLIC ACID, RNA-DEPENDENT RNA-POLYMERASE | | Authors: | Wonacott, A, Skarzynski, T, Singh, O.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of Novel Acyl Pyrrolidine Inhibitors of Hepatitis C Virus RNA-Dependent RNA Polymerase Leading to a Development Candidate.
J.Med.Chem., 50, 2007
|
|
2K8O
 
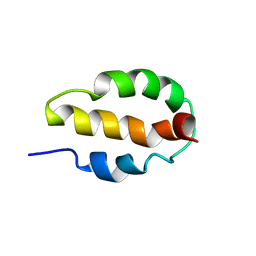 | |
7RX2
 
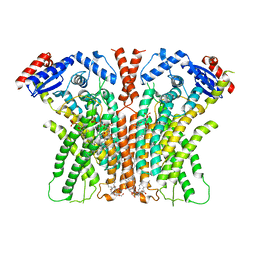 | |
