6LPI
 
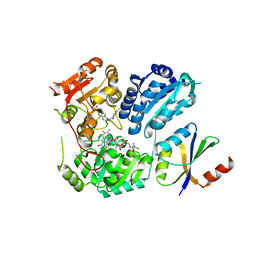 | | Crystal Structure of AHAS holo-enzyme | | Descriptor: | Acetolactate synthase isozyme 1 large subunit, Acetolactate synthase isozyme 1 small subunit, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, Y, Yang, X, Xi, Z, Shen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Molecular architecture of the acetohydroxyacid synthase holoenzyme.
Biochem.J., 477, 2020
|
|
6MG8
 
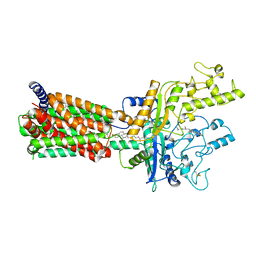 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | Descriptor: | CHOLESTEROL, Protein patched homolog 1 | | Authors: | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
8JY0
 
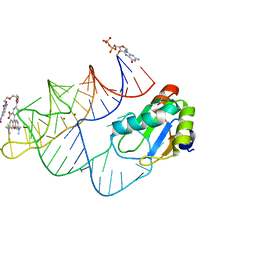 | | Crystal structure of RhoBAST complexed with TMR-DN | | Descriptor: | 2,4-dinitroaniline, 5-aminocarbonyl-2-[3-(dimethylamino)-6-dimethylazaniumylidene-xanthen-9-yl]benzoate, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, Xiao, Y, Xu, Z, Fang, X. | | Deposit date: | 2023-07-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms for binding and activation of a contact-quenched fluorophore by RhoBAST.
Nat Commun, 15, 2024
|
|
6A37
 
 | |
6O7G
 
 | |
2GHQ
 
 | |
2GHT
 
 | |
6UTU
 
 | | Crystal structure of minor pseudopilin ternary complex of XcpVWX from the Type 2 secretion system of Pseudomonas aeruginosa in the P3 space group | | Descriptor: | CALCIUM ION, Type II secretion system protein I, Type II secretion system protein J, ... | | Authors: | Zhang, Y, Wang, S, Jia, Z. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | In Situ Proteolysis Condition-Induced Crystallization of the XcpVWX Complex in Different Lattices.
Int J Mol Sci, 21, 2020
|
|
6VYM
 
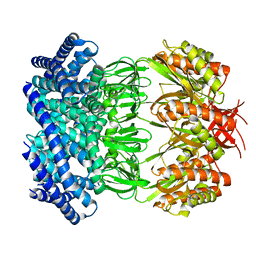 | | Cryo-EM structure of mechanosensitive channel MscS in PC-18:1 nanodiscs treated with beta-cyclodextran | | Descriptor: | Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
6VYL
 
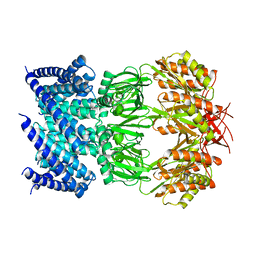 | | Cryo-EM structure of mechanosensitive channel MscS in PC-10 nanodiscs | | Descriptor: | Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
6VRJ
 
 | |
6VYK
 
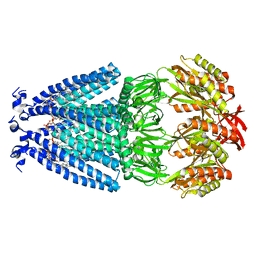 | | Cryo-EM structure of mechanosensitive channel MscS in PC-18:1 nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
6O3W
 
 | |
6O3Y
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM3 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
3TKZ
 
 | | Structure of the SHP-2 N-SH2 domain in a 1:2 complex with RVIpYFVPLNR peptide | | Descriptor: | PROTEIN (RVIpYFVPLNR peptide), Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
3TL0
 
 | | Structure of SHP2 N-SH2 domain in complex with RLNpYAQLWHR peptide | | Descriptor: | RLNpYAQLWHR peptide, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
2MN6
 
 | |
2MN7
 
 | |
2MI2
 
 | |
2O3J
 
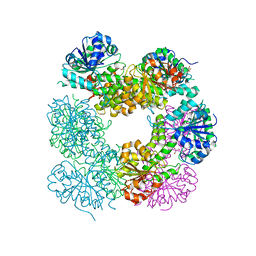 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
2P72
 
 | | crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhang, Y, Xiang, Y, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
2P73
 
 | | crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Y, Xiang, Y, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
7XH3
 
 | |
7XFV
 
 | |
2P6W
 
 | | Crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein | | Authors: | Zhang, Y, Ye, X, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
