2MDK
 
 | |
2N2C
 
 | | NMR Structure of TDP-43 prion-like hydrophobic helix in DPC | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Lim, L, Song, J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ALS-causing mutations significantly perturb the self-assembly and interaction with nucleic acid of the intrinsically-disordered prion-like domain of TDP-43
To be Published
|
|
2MP3
 
 | | Truncated L126Z-sod1 in DPC micelle | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Lim, L, Song, J. | | Deposit date: | 2014-05-10 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism for transforming cytosolic SOD1 into integral membrane proteins of organelles by ALS-causing mutations
Biochim.Biophys.Acta, 1848, 2015
|
|
6KNR
 
 | | Crystal structure of Estrogen-related receptor gamma ligand-binding domain with DN200699 | | Descriptor: | (E)-4-(1-(4-(1-cyclopropylpiperidin-4-yl)phenyl)-5-hydroxy-2-phenylpent-1-en-1-yl)phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Song, J, Cho, S.J. | | Deposit date: | 2019-08-07 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | An orally available inverse agonist of estrogen-related receptor gamma showed expanded efficacy for the radioiodine therapy of poorly differentiated thyroid cancer.
Eur.J.Med.Chem., 205, 2020
|
|
2NAM
 
 | | Full-length WT SOD1 in DPC MICELLE | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Lim, L, Song, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SALS-linked WT-SOD1 adopts a highly similar helical conformation as FALS-causing L126Z-SOD1 in a membrane environment
Biochim.Biophys.Acta, 1858, 2016
|
|
5C6D
 
 | | Crystal structure of USP7 in complex with UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Zhang, Z.-M, Song, J. | | Deposit date: | 2015-06-22 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1.
Cell Rep, 12, 2015
|
|
5EJX
 
 | | X-ray Free Electron Laser Structure of Cytochrome C Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EJT
 
 | | Thermally annealed ferryl Cytochrome C Peroxidase crystal structure | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7DEA
 
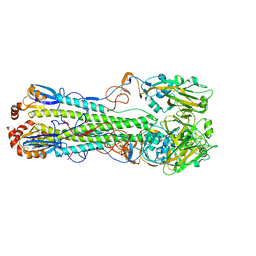 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck Northern China/22/2017 (H5N6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Wei, X, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7DEB
 
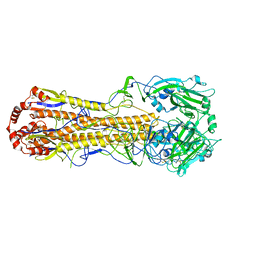 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck/Eastern China/L0230/2010 (H5N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
6A6K
 
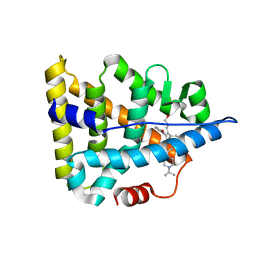 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
5Y70
 
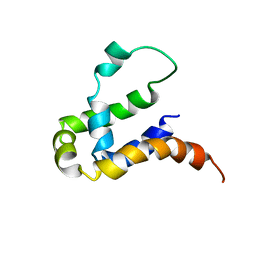 | | NMR structure of KMP11 in DPC micelle | | Descriptor: | Kinetoplastid membrane protein 11 | | Authors: | Lu, Y, Lim, L.Z, Song, J. | | Deposit date: | 2017-08-16 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Kinetoplastid membrane protein-11 adopts a four-helix bundle fold in DPC micelle.
FEBS Lett., 591, 2017
|
|
5XQ5
 
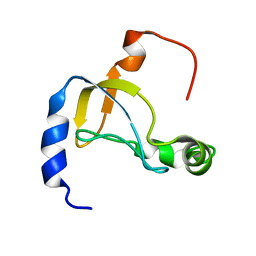 | |
5YX2
 
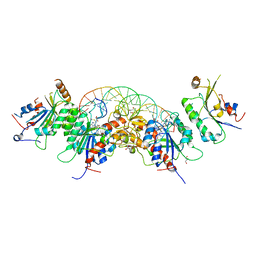 | |
1VJH
 
 | | Crystal structure of gene product of At1g24000 from Arabidopsis thaliana | | Descriptor: | Bet v I allergen family | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1H, 15N and 13C resonance assignments of the putative Bet v 1 family protein At1g24000.1 from Arabidopsis thaliana.
J.Biomol.Nmr, 32, 2005
|
|
7LO8
 
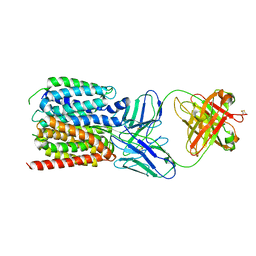 | | NorA in complex with Fab36 | | Descriptor: | Fab36 Heavy Chain, Fab36 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7LO7
 
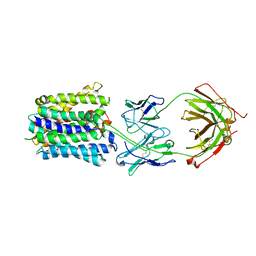 | | NorA in complex with Fab25 | | Descriptor: | Fab25 Heavy Chain, Fab25 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
2QCY
 
 | |
8ZVI
 
 | | Structure of the bacteriophage T5 capsid | | Descriptor: | Decoration protein, Major capsid protein | | Authors: | Peng, Y, Liu, H.R. | | Deposit date: | 2024-06-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection.
Int J Mol Sci, 25, 2024
|
|
8EIH
 
 | | Cryo-EM structure of human DNMT3B homo-tetramer (form I) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIK
 
 | | Cryo-EM structure of human DNMT3B homo-hexamer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIJ
 
 | | Cryo-EM structure of human DNMT3B homo-trimer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EII
 
 | | Cryo-EM structure of human DNMT3B homo-tetramer (form II) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
3GXU
 
 | | Crystal structure of Eph receptor and ephrin complex | | Descriptor: | Ephrin type-A receptor 4, Ephrin-B2 | | Authors: | Qin, H.N, Song, J.X. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the EphA4-ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity
J.Biol.Chem., 2009
|
|
6YEJ
 
 | | Cryo-EM structure of the Full-length disease type human Huntingtin | | Descriptor: | Huntingtin | | Authors: | Tame, G, Jung, T, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2020-03-24 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (18.200001 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
