4YJ6
 
 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | Descriptor: | Aryl acylamidase, PHOSPHATE ION | | Authors: | Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G, Choi, I.-G. | | Deposit date: | 2015-03-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
2GJN
 
 | |
5EJJ
 
 | | Crystal structure of UfSP from C.elegans | | Descriptor: | Ufm1-specific protease | | Authors: | Kim, K, Ha, B, Kim, E.E. | | Deposit date: | 2015-11-02 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Biochem. Biophys. Res. Commun., 476, 2016
|
|
2GJL
 
 | | Crystal Structure of 2-nitropropane dioxygenase | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein PA1024 | | Authors: | Suh, S.W. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 2-Nitropropane Dioxygenase Complexed with FMN and Substrate: identification of the catalytic base
J.Biol.Chem., 281, 2006
|
|
3W8L
 
 | | Crystal structure of human CK2 in complex with inositol hexakisphosphate | | Descriptor: | Casein kinase II subunit alpha, INOSITOL HEXAKISPHOSPHATE | | Authors: | Son, S.H, Lee, W.-K, Yu, Y.G, Lee, H.H. | | Deposit date: | 2013-03-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1QF4
 
 | | DESIGN, SYNTHESIS, AND X-RAY CRYSTAL STRUCTURE OF AN ENZYME BOUND BISUBSTRATE HYBRID INHIBITOR OF ADENYLOSUCCINATE SYNTHETASE | | Descriptor: | (C8-R)-HYDANTOCIDIN 5'-PHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hanessian, S, Lu, P.-P, Sanceau, J.-Y, Chemla, P, Gohda, K, Fonne-Pfister, R, Prade, L, Cowan-Jacob, S.W. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An enzyme-bound bisubstrate hybrid inhibitor of adenylosuccinate synthetase
Angew.Chem.Int.Ed.Engl., 38, 1999
|
|
1QF5
 
 | | DESIGN, SYNTHESIS, AND X-RAY CRYSTAL STRUCTURE OF AN ENZYME BOUND BISUBSTRATE HYBRID INHIBITOR OF ADENYLOSUCCINATE SYNTHETASE | | Descriptor: | (C8-S)-HYDANTOCIDIN 5'-PHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hanessian, S, Lu, P.-P, Sanceau, J.-Y, Chemla, P, Prade, L, Gohda, K, Cowan-Jacob, S.W, Fonne-Pfister, R. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An enzyme-bound bisubstrate hybrid inhibitor of adenylosuccinate synthetase
Angew.Chem.Int.Ed.Engl., 38, 1999
|
|
1RAE
 
 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
7BR9
 
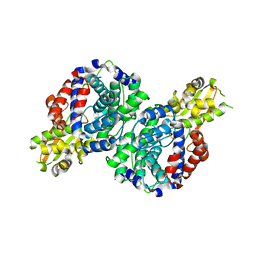 | | Crystal structure of mus musculus IRG1 | | Descriptor: | Cis-aconitate decarboxylase | | Authors: | Park, H.H, Chun, H.L. | | Deposit date: | 2020-03-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of mouse IRG1 suggests that cis-aconitate decarboxylase has an open and closed conformation.
Plos One, 15, 2020
|
|
4RCA
 
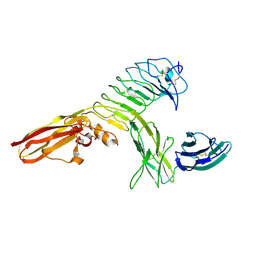 | | Crystal structure of human PTPdelta and human Slitrk1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 1, ... | | Authors: | Kim, H.M, Park, B.S, Kim, D, Lee, S.G. | | Deposit date: | 2014-09-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9908 Å) | | Cite: | Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion.
Nat Commun, 5, 2014
|
|
2OWY
 
 | |
3II5
 
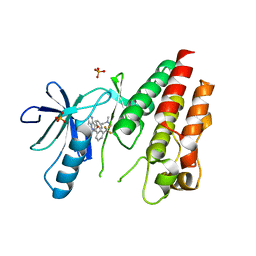 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | Authors: | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2OKL
 
 | |
7DB5
 
 | | Crystal structure of alpha-L-fucosidase from Vibrio sp. strain EJY3 | | Descriptor: | Alpha-L-fucosidase, GLYCEROL, PHOSPHATE ION | | Authors: | Hong, H, Kim, K.-J. | | Deposit date: | 2020-10-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dual alpha-1,4- and beta-1,4-Glycosidase Activities by the Novel Carbohydrate-Binding Module in alpha-l-Fucosidase from Vibrio sp. Strain EJY3.
J.Agric.Food Chem., 69, 2021
|
|
6CDD
 
 | |
1NM3
 
 | | Crystal structure of Heamophilus influenza hybrid-Prx5 | | Descriptor: | Protein HI0572, SULFATE ION | | Authors: | Kim, S.J, Woo, J.R, Hwang, Y.S, Jeong, D.G, Shin, D.H, Kim, K.H, Ryu, S.E. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tetrameric Structure of Haemophilus influenza Hybrid Prx5 Reveals Interactions between Electron Donor and Acceptor Proteins.
J.Biol.Chem., 278, 2003
|
|
6MNY
 
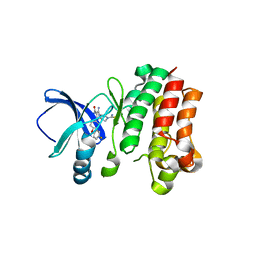 | |
4AT1
 
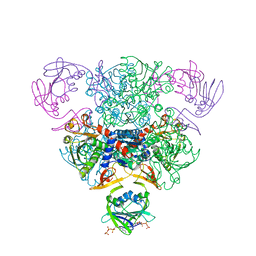 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
1YW8
 
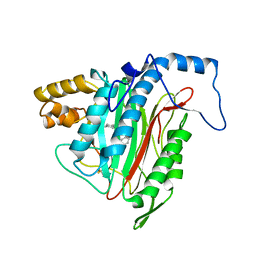 | | h-MetAP2 complexed with A751277 | | Descriptor: | 2-[(PHENYLSULFONYL)AMINO]-5,6,7,8-TETRAHYDRONAPHTHALENE-1-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2005-02-17 | | Release date: | 2006-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
1YW7
 
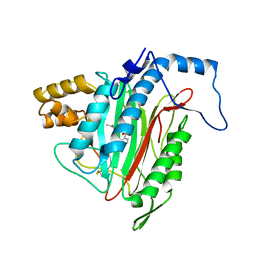 | | h-MetAP2 complexed with A444148 | | Descriptor: | 5-METHYL-2-[(PHENYLSULFONYL)AMINO]BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2005-02-17 | | Release date: | 2006-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
1YW9
 
 | | h-MetAP2 complexed with A849519 | | Descriptor: | 2-[({2-[(1Z)-3-(DIMETHYLAMINO)PROP-1-ENYL]-4-FLUOROPHENYL}SULFONYL)AMINO]-5,6,7,8-TETRAHYDRONAPHTHALENE-1-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2005-02-17 | | Release date: | 2006-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
1AYN
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Oliveira, M.A, Zhao, R, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human rhinovirus 16.
Structure, 1, 1993
|
|
1AYM
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN AT HIGH RESOLUTION | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle.
Structure, 5, 1997
|
|
1C8M
 
 | | REFINED CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS 16 COMPLEXED WITH VP63843 (PLECONARIL), AN ANTI-PICORNAVIRAL DRUG CURRENTLY IN CLINICAL TRIALS | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, HUMAN RHINOVIRUS 16 COAT PROTEIN, ZINC ION | | Authors: | Chakravarty, S, Bator, C.M, Pevear, D.C, Diana, G.D, Rossmann, M.G. | | Deposit date: | 2000-05-26 | | Release date: | 2000-11-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | THE REFINED STRUCTURE OF A PICORNAVIRUS INHIBITOR CURRENTLY IN CLINICAL TRIALS, WHEN COMPLEXED WITH HUMAN RHINOVIRUS 16
to be published
|
|
7AT1
 
 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ... | | Authors: | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | Deposit date: | 1989-09-22 | | Release date: | 1990-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
