8DEG
 
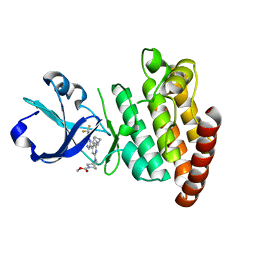 | | Crystal structure of DLK in complex with inhibitor DN0011197 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 12, methyl (1S,4S)-5-{(4P)-4-[5-amino-6-(difluoromethoxy)pyrazin-2-yl]-6-[(1R,4R)-2-azabicyclo[2.1.1]hexan-2-yl]pyridin-2-yl}-2,5-diazabicyclo[2.2.1]heptane-2-carboxylate | | Authors: | Srivastava, A, Lexa, K, de Vicente, J. | | Deposit date: | 2022-06-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of Potent and Selective Dual Leucine Zipper Kinase/Leucine Zipper-Bearing Kinase Inhibitors with Neuroprotective Properties in In Vitro and In Vivo Models of Amyotrophic Lateral Sclerosis.
J.Med.Chem., 65, 2022
|
|
2Y1R
 
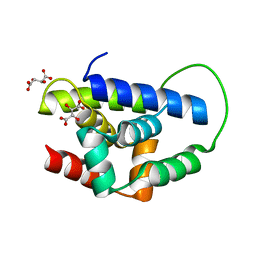 | | Structure of MecA121 & ClpC N-domain complex | | Descriptor: | ADAPTER PROTEIN MECA 1, NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
6BPL
 
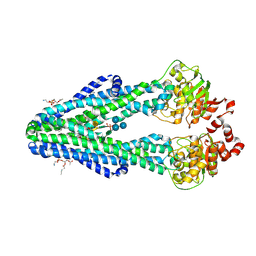 | | E. coli MsbA in complex with LPS and inhibitor G907 | | Descriptor: | (2E)-3-{6-[(1S)-1-(2-chloro-6-cyclopropylphenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-amino-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-23 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
6BPP
 
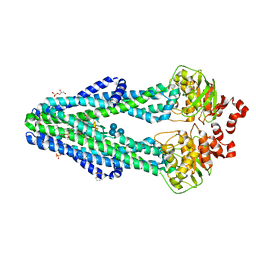 | | E. coli MsbA in complex with LPS and inhibitor G092 | | Descriptor: | (2E)-3-{6-[(1S)-1-(3-amino-2,6-dichlorophenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
8E9F
 
 | |
3JCO
 
 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6DKJ
 
 | | human GIPR ECD and Fab complex | | Descriptor: | 1,2-ETHANEDIOL, Fab heavy chain, Fab light chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models.
Sci Transl Med, 10, 2018
|
|
6X2J
 
 | | Structure of human TRPA1 in complex with agonist GNE551 | | Descriptor: | 5-amino-1-[(4-bromo-2-fluorophenyl)methyl]-N-(2,5-dimethoxyphenyl)-1H-1,2,3-triazole-4-carboxamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Chen, H. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A Non-covalent Ligand Reveals Biased Agonism of the TRPA1 Ion Channel.
Neuron, 109, 2021
|
|
3R4N
 
 | | Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 4-[2-chloro-6-(4,4,4-trifluorobutoxy)phenyl]-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
3R4O
 
 | | Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 2-amino-N-cyclobutyl-4-[2,4-dichloro-6-(4,4,4-trifluorobutoxy)phenyl]-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
6LSA
 
 | | Complex structure of bovine herpesvirus 1 glycoprotein D and bovine nectin-1 IgV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6DY7
 
 | | WDR5 in complex with a WIN site inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N-[3-(2,4-dichlorophenoxy)propyl]-1H-imidazol-2-amine, SULFATE ION, ... | | Authors: | Phan, J, Wang, F, Fesik, S.W. | | Deposit date: | 2018-07-01 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity.
Cell Rep, 26, 2019
|
|
6DYA
 
 | | WDR5 in complex with a WIN site inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3,5-dichlorophenyl)methyl]-3-[(1H-imidazol-1-yl)methyl]benzamide, SULFATE ION, ... | | Authors: | Phan, J, Wang, F, Fesik, S.W. | | Deposit date: | 2018-07-01 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity.
Cell Rep, 26, 2019
|
|
3R4P
 
 | | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 2-amino-4-{2,4-dichloro-6-[2-(1H-pyrazol-1-yl)ethoxy]phenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
3R4M
 
 | | Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 4-CHLORO-6-(2-METHOXYPHENYL)PYRIMIDIN-2-AMINE, Heat shock protein HSP 90-alpha | | Authors: | Almassy, R.J. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
2Y1Q
 
 | | Crystal Structure of ClpC N-terminal Domain | | Descriptor: | NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, SULFATE ION | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
3NGO
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | Descriptor: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGQ
 
 | | Crystal structure of the human CNOT6L nuclease domain | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
4M38
 
 | |
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
6WI7
 
 | | RING1B-BMI1 fusion in closed conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1 chimera, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
6WI8
 
 | |
3KPX
 
 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
6M79
 
 | |
