8TLM
 
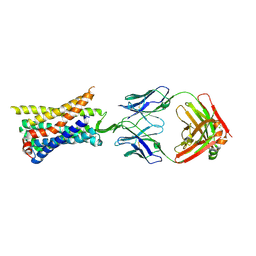 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8EL0
 
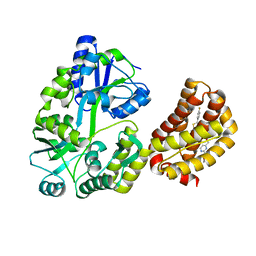 | | Structure of MBP-Mcl-1 in complex with a macrocyclic compound | | Descriptor: | (7R,20P)-18-chloro-1-(4-fluorophenyl)-10-{[(2M)-2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy}-19-methyl-15-[2-(4-methylpiperazin-1-yl)ethyl]-7,8,15,16-tetrahydro-14H-17,20-etheno-9,13-(metheno)-6-oxa-2-thia-3,5,15-triazacyclooctadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
8EL1
 
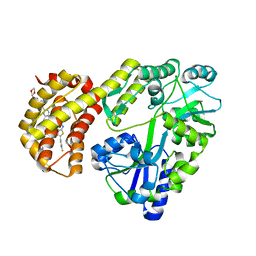 | | Structure of MBP-Mcl-1 in complex with ABBV-467 | | Descriptor: | (7R,16R)-19,23-dichloro-10-{[2-(4-{[(2R)-1,4-dioxan-2-yl]methoxy}phenyl)pyrimidin-4-yl]methoxy}-1-(4-fluorophenyl)-20,22-dimethyl-16-[(4-methylpiperazin-1-yl)methyl]-7,8,15,16-tetrahydro-18,21-etheno-13,9-(metheno)-6,14,17-trioxa-2-thia-3,5-diazacyclononadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
7CMZ
 
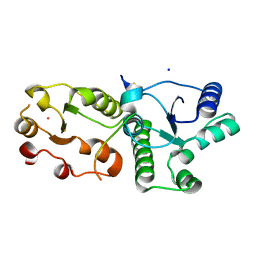 | | Crystal Structure of BRCT7/8 in Complex with the APS Motif of PHF8 | | Descriptor: | DNA topoisomerase 2-binding protein 1, Histone lysine demethylase PHF8, POTASSIUM ION, ... | | Authors: | Che, S.Y, Ma, S, Cao, C, Yao, Z, Shi, L, Yang, N. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability.
Sci Adv, 7, 2021
|
|
6M79
 
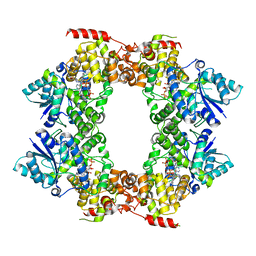 | |
8DEG
 
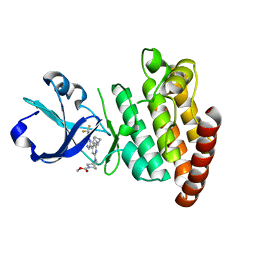 | | Crystal structure of DLK in complex with inhibitor DN0011197 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 12, methyl (1S,4S)-5-{(4P)-4-[5-amino-6-(difluoromethoxy)pyrazin-2-yl]-6-[(1R,4R)-2-azabicyclo[2.1.1]hexan-2-yl]pyridin-2-yl}-2,5-diazabicyclo[2.2.1]heptane-2-carboxylate | | Authors: | Srivastava, A, Lexa, K, de Vicente, J. | | Deposit date: | 2022-06-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of Potent and Selective Dual Leucine Zipper Kinase/Leucine Zipper-Bearing Kinase Inhibitors with Neuroprotective Properties in In Vitro and In Vivo Models of Amyotrophic Lateral Sclerosis.
J.Med.Chem., 65, 2022
|
|
4S3G
 
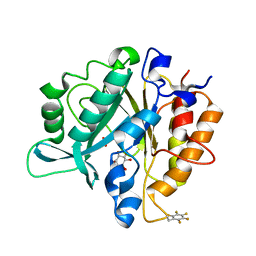 | | Structure of the F249X mutant of Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | He, T, Gershenson, A, Eyles, S.J, Gao, J, Roberts, M.F. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-01 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fluorinated Aromatic Amino Acids Distinguish Cation-pi Interactions from Membrane Insertion.
J.Biol.Chem., 290, 2015
|
|
6VQF
 
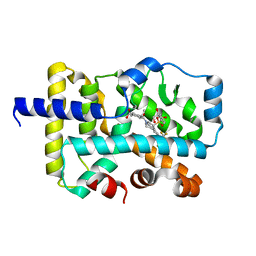 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244- 487)-L6-SRC1(678-692)) IN COMPLEX WITH AN INVERSE AGONIST | | Descriptor: | (1R,3S,4R)-4-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]-3-methylcyclohexane-1-carboxylic acid, GLYCEROL, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-02-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of BMS-986251: A Clinically Viable, Potent, and Selective ROR gamma t Inverse Agonist.
Acs Med.Chem.Lett., 11, 2020
|
|
4ZNO
 
 | | Crystal structure of Dln1 complexed with sucrose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Natterin-like protein, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4ZNQ
 
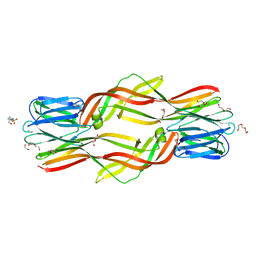 | | Crystal structure of Dln1 complexed with Man(alpha1-2)Man | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4ZNR
 
 | | Crystal structure of Dln1 complexed with Man(alpha1-3)Man | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4RV3
 
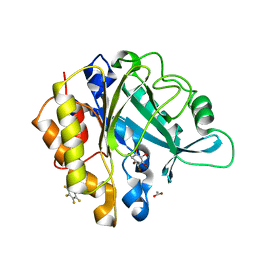 | | Crystal structure of a pentafluoro-Phe incorporated Phosphatidylinositol-specific phospholipase C (H258X)from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | He, T, Gershenson, A, Eyles, S.J, Gao, J, Roberts, M.F. | | Deposit date: | 2014-11-24 | | Release date: | 2015-07-01 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fluorinated Aromatic Amino Acids Distinguish Cation-pi Interactions from Membrane Insertion.
J.Biol.Chem., 290, 2015
|
|
6BPL
 
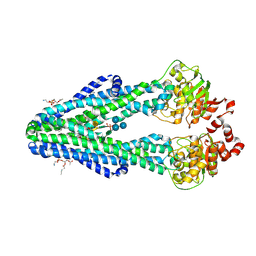 | | E. coli MsbA in complex with LPS and inhibitor G907 | | Descriptor: | (2E)-3-{6-[(1S)-1-(2-chloro-6-cyclopropylphenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-amino-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-23 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
6U25
 
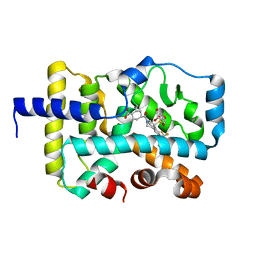 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS- RORGT(244-487)-L6-SRC1(678-692)) IN COMPLEX WITH A TRICYCLIC INVERSE AGONIST | | Descriptor: | GLYCEROL, NUCLEAR RECEPTOR COACTIVATOR 1 CHIMERA, trans-4-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]cyclohexane-1-carboxylic acid | | Authors: | Sack, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Rationally Designed, Conformationally Constrained Inverse Agonists of ROR gamma t-Identification of a Potent, Selective Series with Biologic-Like in Vivo Efficacy.
J.Med.Chem., 62, 2019
|
|
1L0Q
 
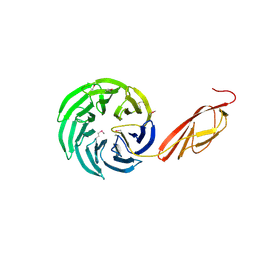 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
7C8D
 
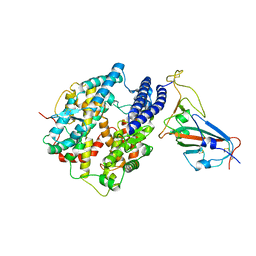 | | Cryo-EM structure of cat ACE2 and SARS-CoV-2 RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1, ZINC ION | | Authors: | Gao, G.F, Wang, Q.H, Wu, L.l. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2.
Cell Discov, 6, 2020
|
|
8HKY
 
 | |
8HKX
 
 | |
8HKZ
 
 | |
8HKU
 
 | |
8HKV
 
 | |
4D90
 
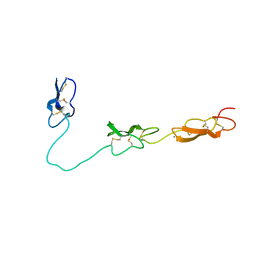 | | Crystal Structure of Del-1 EGF domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q, Schurpf, T, Springer, T, Wang, J. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The RGD finger of Del-1 is a unique structural feature critical for integrin binding.
Faseb J., 26, 2012
|
|
8HL2
 
 | |
8HL5
 
 | |
8HL3
 
 | |
