8PXV
 
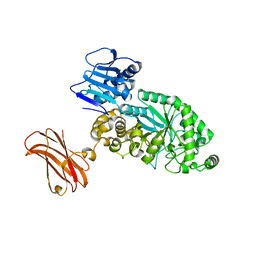 | | Targeting extended blood antigens by Akkermansia muciniphila enzymes unveils a missing link for generating universal donor blood | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL, SODIUM ION, ... | | Authors: | Weikum, J, Jensen, M, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Akkermansia muciniphila exoglycosidases target extended blood group antigens to generate ABO-universal blood.
Nat Microbiol, 9, 2024
|
|
8PXU
 
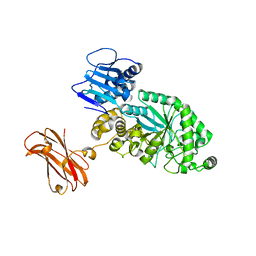 | |
8PXT
 
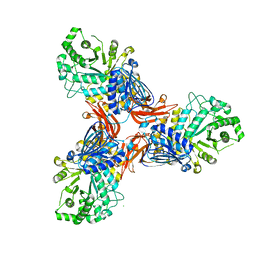 | | Targeting extended blood antigens by Akkermansia muciniphila enzymes unveils a missing link for generating universal donor blood | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Jensen, M, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Akkermansia muciniphila exoglycosidases target extended blood group antigens to generate ABO-universal blood.
Nat Microbiol, 9, 2024
|
|
7UK2
 
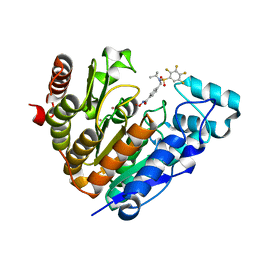 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with NN-390 | | Descriptor: | Hdac6 protein, N-hydroxy-4-{[(propan-2-yl)(2,3,4,5-tetrafluorobenzene-1-sulfonyl)amino]methyl}benzamide, POTASSIUM ION, ... | | Authors: | Erdogan, F, Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Efficacy and Drug Synergy of HDAC6-Selective Inhibitor NN-429 in Natural Killer (NK)/T-Cell Lymphoma.
Pharmaceuticals, 15, 2022
|
|
6BQ8
 
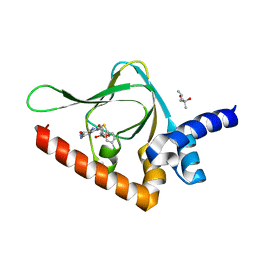 | | Joint X-ray/neutron structure of PKG II CNB-B domain in complex with 8-pCPT-cGMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(~2~H_2_)amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2-hydroxy-7-(~2~H)hydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl](~2~H)-1,9-dihydro-6H-purin-6-one, STRONTIUM ION, ... | | Authors: | Kim, C, Kovalevsky, A, Gerlits, O. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-21 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Neutron Crystallography Detects Differences in Protein Dynamics: Structure of the PKG II Cyclic Nucleotide Binding Domain in Complex with an Activator.
Biochemistry, 57, 2018
|
|
4YE7
 
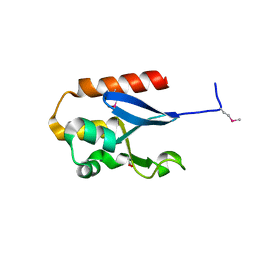 | |
8QZO
 
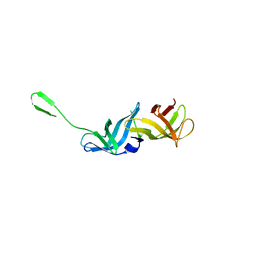 | |
8OT2
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-5-methyl-phenol, ACETATE ION, ... | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
8F43
 
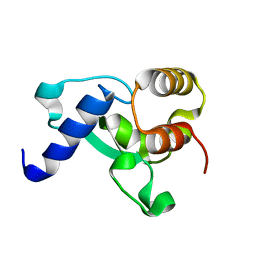 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | Descriptor: | CRISPR-associated endonuclease Cas9 | | Authors: | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
8FSD
 
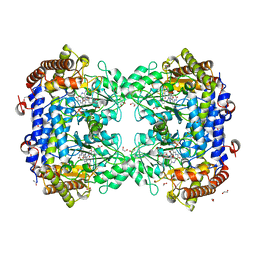 | | P130R mutant of soybean SHMT8 in complex with PLP-glycine and formylTHF | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Beamer, L.J, Korasick, D.A. | | Deposit date: | 2023-01-09 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional analysis of two SHMT8 variants associated with soybean cyst nematode resistance.
Febs J., 291, 2024
|
|
6TX4
 
 | |
6TX7
 
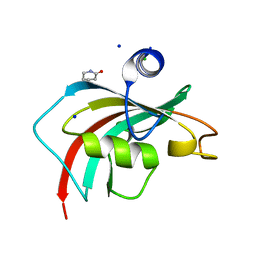 | |
6TX5
 
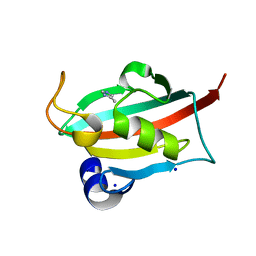 | |
8GAR
 
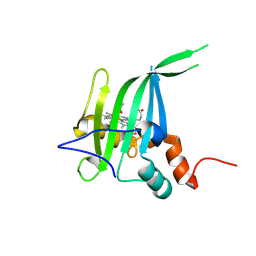 | |
6MF0
 
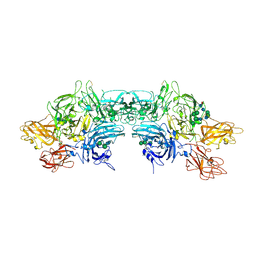 | | Crystal Structure Determination of Human/Porcine Chimera Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.19999886 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
6MRV
 
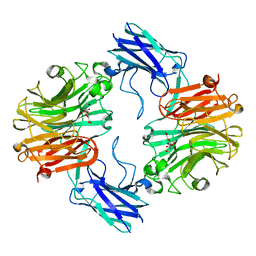 | | Sialidase26 co-crystallized with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MF2
 
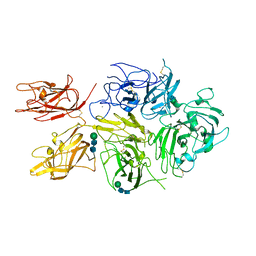 | | Improved Model of Human Coagulation Factor VIII | | Descriptor: | CALCIUM ION, COPPER (I) ION, Coagulation factor VIII, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.609364 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
6MRX
 
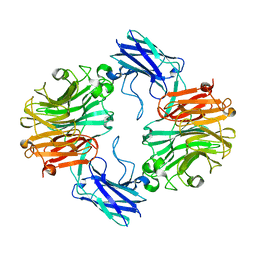 | | Sialidase26 apo | | Descriptor: | Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MYV
 
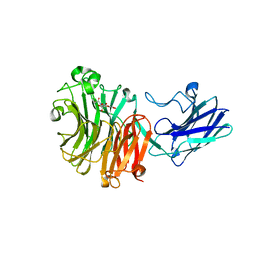 | | Sialidase26 co-crystallized with DANA-Gc | | Descriptor: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MNJ
 
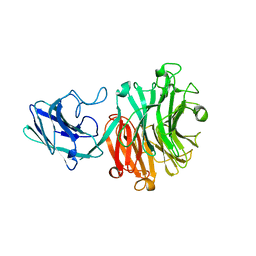 | | Hadza microbial sialidase Hz136 | | Descriptor: | Hz136 | | Authors: | Rees, S.D, Zaramela, L, Zengler, K, Chang, G. | | Deposit date: | 2018-10-01 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6VIF
 
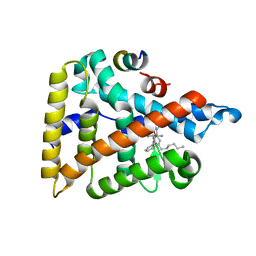 | | Human LRH-1 ligand-binding domain bound to agonist cpd 15 and fragment of coregulator TIF-2 | | Descriptor: | N-[(8beta,11alpha,12alpha)-8-{[methyl(phenyl)amino]methyl}-1,6:7,14-dicycloprosta-1(6),2,4,7(14)-tetraen-11-yl]sulfuric diamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Development of a new class of liver receptor homolog-1 (LRH-1) agonists by photoredox conjugate addition.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WYH
 
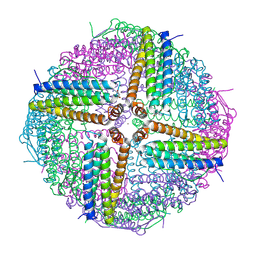 | |
7T69
 
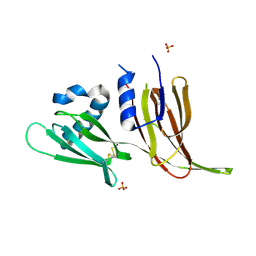 | | Crystal structure of Avr3 (SIX1) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr3 (SIX1), Secreted in xylem 1, SULFATE ION | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
7T6A
 
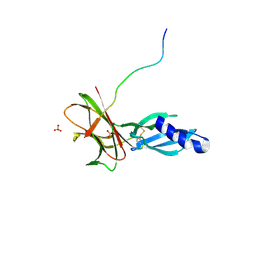 | | Crystal structure of Avr1 (SIX4) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr1 (FolSIX4), Avirulence protein 1, Avr1 (SIX4), ... | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
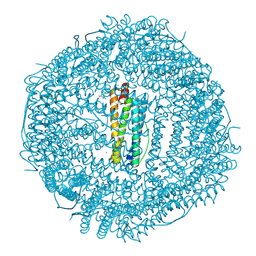
6WYF
 
 | |
