9QB4
 
 | |
8RHJ
 
 | | Yeast 20S proteasome in complex with a macrocyclic oxindole epoxyketone (compound 5) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Macrocyclic oxindole epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
8RHL
 
 | | Yeast 20S proteasome in complex with a linear biarylether epoxyketone (compound 15a) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Linear biarylether epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
8RHK
 
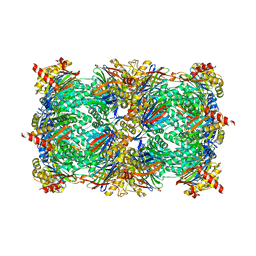 | | Yeast 20S proteasome in complex with a linear oxindole epoxyketone (compound 6) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Linear oxindole epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
6EK8
 
 | | YaxB from Yersinia enterocolitica | | Descriptor: | YaxB | | Authors: | Braeuning, B, Groll, M. | | Deposit date: | 2017-09-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB.
Nat Commun, 9, 2018
|
|
6TTY
 
 | | Structure of ClpP from Staphylococcus aureus (apo, closed state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
6TTZ
 
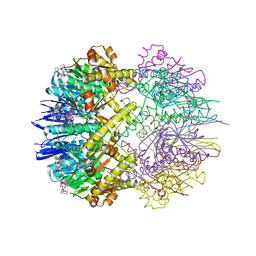 | | Structure of the ClpP:ADEP4-complex from Staphylococcus aureus (open state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, N-[(2S)-3-(3,5-difluorophenyl)-1-[[(3S,9S,13S,15R,19S,22S)-15,19-dimethyl-2,8,12,18,21-pentaoxo-11-oxa-1,7,17,20-tetrazatetracyclo[20.4.0.03,7.013,17]hexacosan-9-yl]amino]-1-oxopropan-2-yl]heptanamide | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
4BA8
 
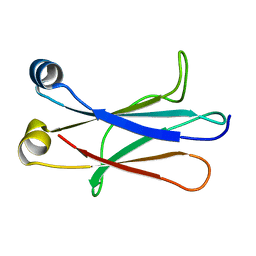 | | High resolution NMR structure of the C mu3 domain from IgM | | Descriptor: | IG MU CHAIN C REGION SECRETED FORM | | Authors: | Mueller, R, Kern, T, Graewert, M.A, Madl, T, Peschek, J, Groll, M, Sattler, M, Buchner, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-06-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High Resolution Structures of the Igm Fc Domains Reveal Principles of its Hexamer Formation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8OLL
 
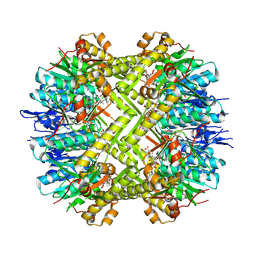 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OLR
 
 | | Structure of yeast 20S proteasome in complex with the natural product beta-lactone inhibitor Cystargolide A | | Descriptor: | CHLORIDE ION, Cystargolide A (bound), MAGNESIUM ION, ... | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8A82
 
 | | Fe(II)/aKG-dependent halogenase OocPQ | | Descriptor: | Cupin_8 domain-containing protein, FE (III) ION, GLYCEROL, ... | | Authors: | Fraley, A.E, Meoded, R.A, Schmalhofer, M, Bergande, C, Groll, M, Piel, J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Heterocomplex structure of a polyketide synthase component involved in modular backbone halogenation.
Structure, 31, 2023
|
|
7ZHD
 
 | | Crystal structure of CtaZ in complex with Closthioamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, Transcription activator effector binding, ... | | Authors: | Gude, F, Molloy, E.M, Horch, T, Dell, M, Dunbar, K.L, Krabbe, J, Groll, M, Hertweck, C. | | Deposit date: | 2022-04-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Specialized Polythioamide-Binding Protein Confers Antibiotic Self-Resistance in Anaerobic Bacteria.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7ZHE
 
 | | Crystal structure of CtaZ from Ruminiclostridium cellulolyticum | | Descriptor: | GLYCEROL, SODIUM ION, Transcription activator effector binding | | Authors: | Gude, F, Molloy, E.M, Horch, T, Dell, M, Dunbar, K.L, Krabbe, J, Groll, M, Hertweck, C. | | Deposit date: | 2022-04-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Specialized Polythioamide-Binding Protein Confers Antibiotic Self-Resistance in Anaerobic Bacteria.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7ZEI
 
 | | Thermostable GH159 glycoside hydrolase from Caldicellulosiruptor at 1.7 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Baudrexl, M, Fida, T, Berk, B, Schwarz, W, Zverlov, V.V, Groll, M, Liebl, W. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of Thermostable GH159 Glycoside Hydrolases Exhibiting alpha-L-Arabinofuranosidase Activity.
Front Mol Biosci, 9, 2022
|
|
5JHR
 
 | | Yeast 20S proteasome in complex with the peptidic epoxyketone inhibitor 27 | | Descriptor: | (2S)-2-azido-N-[(2S)-3-(biphenyl-4-yl)-1-{[(2S)-1-{[(2S,3S,4R)-3,5-dihydroxy-4-methylpentan-2-yl]amino}-1-oxo-3-phenylpropan-2-yl]amino}-1-oxopropan-2-yl]-3-phenylpropanamide (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-04-21 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of beta 5c Selective Inhibitors of Human Constitutive Proteasomes.
J.Med.Chem., 59, 2016
|
|
5JHS
 
 | |
5YVR
 
 | | Crystal Structure of the H277A mutant of ADH/D1, an archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
5YVM
 
 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NZQ | | Descriptor: | 5,6-DIHYDROXY-NADP, MANGANESE (II) ION, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-26 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
5YVS
 
 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NADP | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
8RDO
 
 | |
8RDM
 
 | | Holomycin methyltransferase DtpM with SAH | | Descriptor: | DtpM, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isofunctional but Structurally Different Methyltransferases for Dithiolopyrrolone Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RDN
 
 | | Holomycin methyltransferase DtpM with SAH and XRD-271 | | Descriptor: | CHLORIDE ION, DtpM, GLYCEROL, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isofunctional but Structurally Different Methyltransferases for Dithiolopyrrolone Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RDL
 
 | | Xenorhabdin methyltransferase XrdM with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ToxA protein | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isofunctional but Structurally Different Methyltransferases for Dithiolopyrrolone Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5CZA
 
 | | Yeast 20S proteasome beta5-D166N mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5CZ9
 
 | |
