6BLK
 
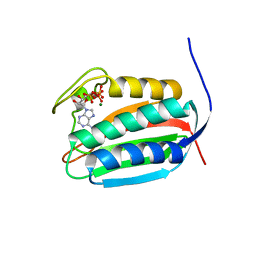 | | Mycobacterial sensor histidine kinase MprB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Signal transduction histidine-protein kinase/phosphatase mprB | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2017-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Mycobacterial sensor histidine kinase MprB
to be published
|
|
7W1Y
 
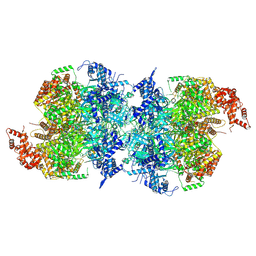 | | Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (49-MER), ... | | Authors: | Li, J, Dong, J, Dang, S, Zhai, Y. | | Deposit date: | 2021-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | The human pre-replication complex is an open complex.
Cell, 186, 2023
|
|
8W6I
 
 | |
8W6J
 
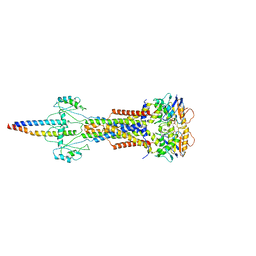 | | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, He, Y, Luo, M. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc
To Be Published
|
|
4JOD
 
 | |
4JOB
 
 | |
4JOC
 
 | |
8YCM
 
 | | Monomeric Human STK19 | | Descriptor: | Isoform 4 of Inactive serine/threonine-protein kinase 19, SULFATE ION | | Authors: | Li, J, Ma, X, Dong, Z. | | Deposit date: | 2024-02-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Mutations found in cancer patients compromise DNA binding of the winged helix protein STK19.
Sci Rep, 14, 2024
|
|
8UWU
 
 | | EmrE structure in the proton-bound state (WT/L51I heterodimer) | | Descriptor: | SMR family multidrug efflux protein EmrE | | Authors: | Li, J, Sae Her, A, Besch, A, Ramirez, B, Crames, M, Banigan, J.R, Mueller, C, Marsiglia, W.M, Zhang, Y, Traaseth, N.J. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Nat Commun, 15, 2024
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
5XBF
 
 | | Crystal Structure of Myo7b C-terminal MyTH4-FERM in complex with USH1C PDZ3 | | Descriptor: | ACETATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Li, J, He, Y, Weck, W.L, Lu, Q, Tyska, M.J, Zhang, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XR9
 
 | | Crystal structure of Human Hsp90 with FS6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-bromanyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of Human Hsp90 with FS6
To Be Published
|
|
5XQE
 
 | | Crystal structure of Human Hsp90 with FS3 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2-methyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of Human Hsp90 with FS3
To Be Published
|
|
5XRD
 
 | |
5XQD
 
 | | Crystal structure of Human Hsp90 with FS2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human Hsp90 with FS2
To Be Published
|
|
5XR5
 
 | | Crystal structure of Human Hsp90 with FS4 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2,2-dimethyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human Hsp90 with FS4
To Be Published
|
|
5XRB
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5
To Be Published
|
|
8HHE
 
 | |
5YIS
 
 | | Crystal Structure of AnkB LIR/LC3B complex | | Descriptor: | Ankyrin-2, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIQ
 
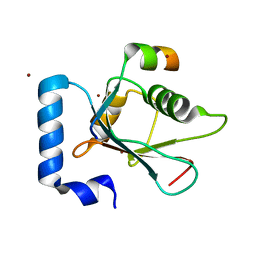 | | Crystal structure of AnkG LIR/LC3B complex | | Descriptor: | Ankyrin-3, Microtubule-associated proteins 1A/1B light chain 3B, ZINC ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIR
 
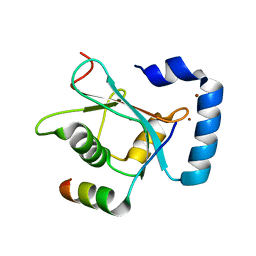 | | Crystal Structure of AnkB LIR/GABARAP complex | | Descriptor: | Ankyrin-2, Gamma-aminobutyric acid receptor-associated protein, NICKEL (II) ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIP
 
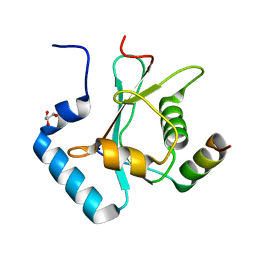 | | Crystal Structure of AnkG LIR/GABARAPL1 complex | | Descriptor: | Ankyrin-3, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5WST
 
 | | Crystal structure of Myo7a SAH | | Descriptor: | Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
5XRE
 
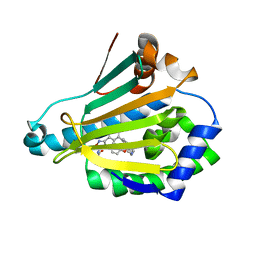 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[4-(4-methoxyphenyl)-3-[5-(8-methylquinolin-5-yl)-2,4-bis(oxidanyl)phenyl]-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1
To Be Published
|
|
