8JJ2
 
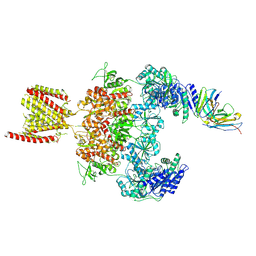 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in one fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 2024
|
|
5YKN
 
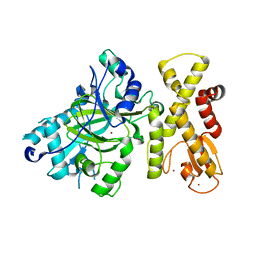 | | crystal structure of Arabidopsis thaliana JMJ14 catalytic domain | | Descriptor: | NICKEL (II) ION, Probable lysine-specific demethylase JMJ14, ZINC ION | | Authors: | Yang, Z, Du, J. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases.
Plant Cell, 30, 2018
|
|
8WG7
 
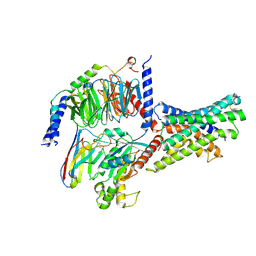 | | Cryo-EM structures of peptide free and Gs-coupled GLP-1R | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8WG8
 
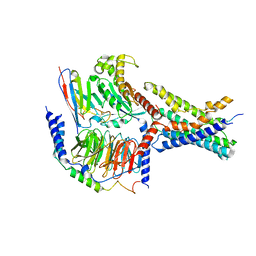 | | Cryo-EM structures of peptide free and Gs-coupled GCGR | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
5IC7
 
 | | Structure of the WD domain of UTP18 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.331 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
8X02
 
 | | E2 core of 2-oxoglutarate dehydrogenase complex | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Zhang, S.S, Zhang, Y.T. | | Deposit date: | 2023-11-03 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular architecture of the mammalian 2-oxoglutarate dehydrogenase complex.
Nat Commun, 15, 2024
|
|
5ICA
 
 | | Structure of the CTD complex of UTP12, Utp13, Utp1 and Utp21 | | Descriptor: | Periodic tryptophan protein 2-like protein, Putative U3 snoRNP protein, Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.507 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC9
 
 | | Structure of the CTD complex of Utp12 and Utp13 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC8
 
 | | Structure of UTP6 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
6KHY
 
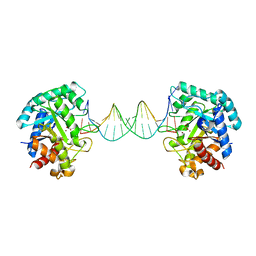 | | The crystal structure of AsfvAP:AG | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (AGCGTCACCGACGAGGC), DNA(AGCGTCACCGACGAGG), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
3HYF
 
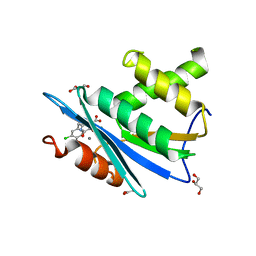 | | Crystal structure of HIV-1 RNase H p15 with engineered E. coli loop and active site inhibitor | | Descriptor: | 2-(3,4-dichlorobenzyl)-5,6-dihydroxypyrimidine-4-carboxylic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Lansdon, E.B, Kirschberg, T.A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information.
J.Med.Chem., 52, 2009
|
|
7WCT
 
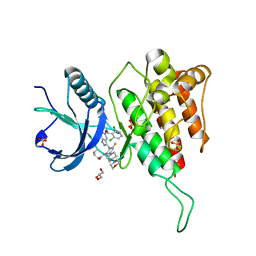 | | Crystal structure of FGFR4 kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7WCW
 
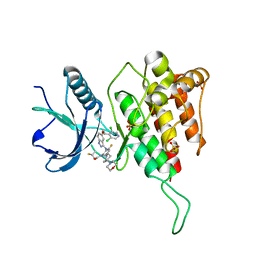 | | Crystal structure of FGFR4(V550L) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.317 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7WCX
 
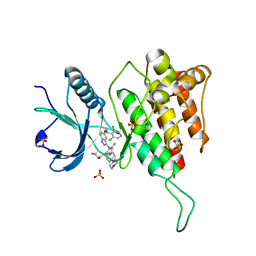 | | Crystal structure of FGFR4(V550M) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7YBO
 
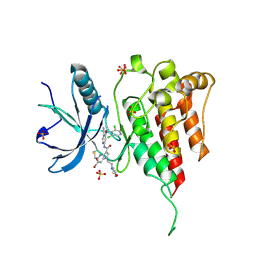 | | Crystal structure of FGFR4 kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC3
 
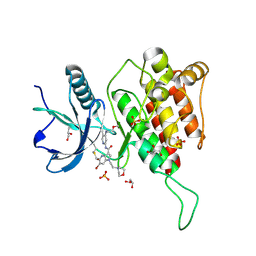 | | Crystal structure of FGFR4 kinase domain with 10t | | Descriptor: | 6-bromanyl-~{N}-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide, Fibroblast growth factor receptor 4, GLYCEROL, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC1
 
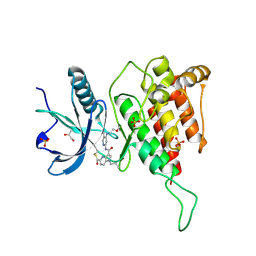 | | Crystal structure of FGFR4 kinase domain with 10d | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YBP
 
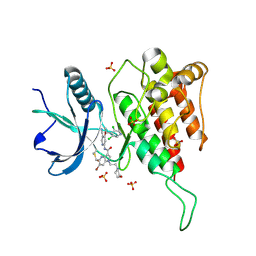 | | Crystal structure of FGFR4(V550L) kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YBX
 
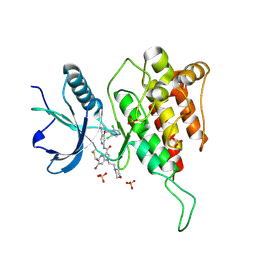 | | Crystal structure of FGFR4(V550M) kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
6LJL
 
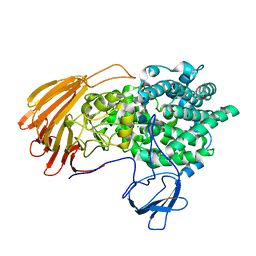 | | Crystal Structure of exoHep-Y390A/H555A complexed with a tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
5C93
 
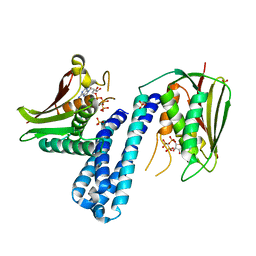 | | Histidine kinase with ATP | | Descriptor: | Histidine kinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SULFATE ION | | Authors: | Cai, Y. | | Deposit date: | 2015-06-26 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6N3H
 
 | | Crystal structure of Kelch domain of the human NS1 binding protein | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B.M.A, Chook, Y.M. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DWO
 
 | |
6N34
 
 | | Crystal structure of the BTB domain of Human NS1-BP | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B, Chook, Y.M. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8JZX
 
 | | SLC15A4 inhibitor complex | | Descriptor: | 2-(4-ethoxyphenyl)-N-[3-[(2R)-2-methylpiperidin-1-yl]propyl]quinoline-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Zhang, S.S, Chen, X.D, Xie, M. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A conformation-locking inhibitor of SLC15A4 with TASL proteostatic anti-inflammatory activity.
Nat Commun, 14, 2023
|
|
