8ZSJ
 
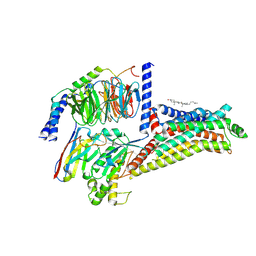 | | Cryo-EM structure of the apo hTAAR1-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
4MY2
 
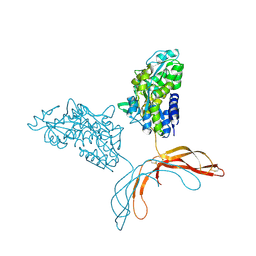 | | Crystal Structure of Norrin in fusion with Maltose Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, Norrin fusion protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ke, J, Jurecky, C, Chen, C, Gu, X, Parker, N, Williams, B.O, Melcher, K, Xu, H.E. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function of Norrin in assembly and activation of a Frizzled 4-Lrp5/6 complex.
Genes Dev., 27, 2013
|
|
7XP6
 
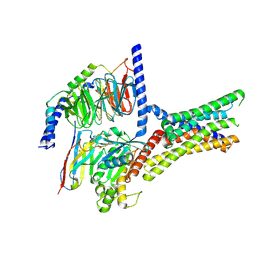 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
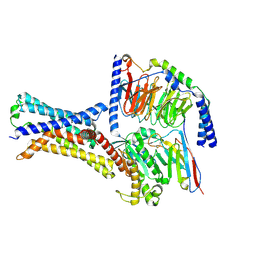 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP5
 
 | | Cryo-EM structure of a class T GPCR in ligand-free state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XDI
 
 | | Tail structure of bacteriophage SSV19 | | Descriptor: | B210, C131, VP1, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2022-03-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into a spindle-shaped archaeal virus with a sevenfold symmetrical tail.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YJM
 
 | | Cryo-EM structure of the monomeric atSPT-ORM1 complex | | Descriptor: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJN
 
 | | Cryo-EM structure of the monomeric atSPT-ORM1 (ORM1-N17A) complex | | Descriptor: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJO
 
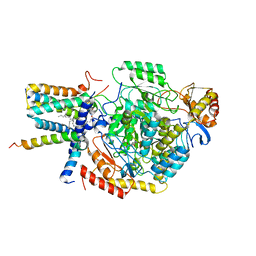 | | Cryo-EM structure of the monomeric atSPT-ORM1 (LCB2a-deltaN5) complex | | Descriptor: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJK
 
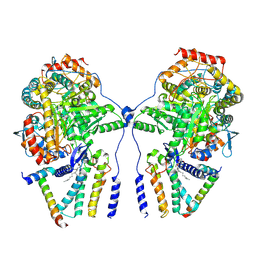 | | Cryo-EM structure of the dimeric atSPT-ORM1 complex | | Descriptor: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
8U56
 
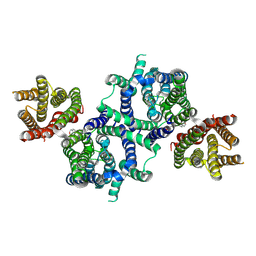 | |
8U5C
 
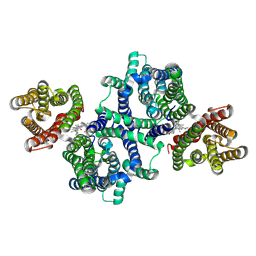 | |
8U54
 
 | |
8U5V
 
 | |
8U5X
 
 | |
8U58
 
 | |
8U5N
 
 | |
8U5Q
 
 | |
6LRD
 
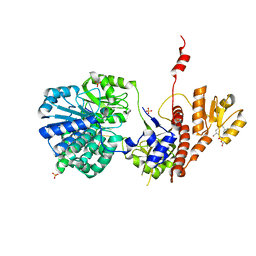 | | Structure of RecJ complexed with a 5'-P-dSpacer-modified ssDNA | | Descriptor: | ASP-LEU-PRO-PHE, DNA (5'-D(P*(3DR)P*TP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Cheng, K, Hua, Y. | | Deposit date: | 2020-01-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901335 Å) | | Cite: | Participation of RecJ in the base excision repair pathway of Deinococcus radiodurans.
Nucleic Acids Res., 48, 2020
|
|
3GT9
 
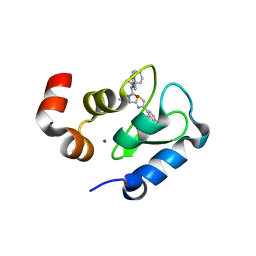 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | Baculoviral IAP repeat-containing 7, N-{(1S)-1-cyclohexyl-2-[(2S)-2-(4-naphthalen-1-yl-1,3-thiazol-2-yl)pyrrolidin-1-yl]-2-oxoethyl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5XS2
 
 | |
5XWW
 
 | |
3GTA
 
 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing 7, ... | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7C6P
 
 | | Bromodomain-containing 4 BD2 in complex with 3',4',7,8- Tetrahydroxyflavonoid | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4 | | Authors: | Li, J, Yu, K, Luo, Y, Zheng, W, Liang, W, Zhu, J. | | Deposit date: | 2020-05-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7C2Z
 
 | | Bromodomain-containing 4 BD1 in complex with 3',4',7,8-Tetrahydroxyflavone | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Li, J, Zhu, J. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
