3D9V
 
 | |
1XAS
 
 | | CRYSTAL STRUCTURE, AT 2.6 ANGSTROMS RESOLUTION, OF THE STREPTOMYCES LIVIDANS XYLANASE A, A MEMBER OF THE F FAMILY OF BETA-1,4-D-GLYCANSES | | Descriptor: | 1,4-BETA-D-XYLAN XYLANOHYDROLASE | | Authors: | Derewenda, U, Derewenda, Z.S. | | Deposit date: | 1994-05-31 | | Release date: | 1995-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure, at 2.6-A resolution, of the Streptomyces lividans xylanase A, a member of the F family of beta-1,4-D-glycanases.
J.Biol.Chem., 269, 1994
|
|
1BWQ
 
 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
1BWP
 
 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
1BWR
 
 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
1JFR
 
 | |
1ES9
 
 | | X-RAY CRYSTAL STRUCTURE OF R22K MUTANT OF THE MAMMALIAN BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASES (PAF-AH) | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB GAMMA SUBUNIT | | Authors: | McMullen, T.W.P, Li, J, Sheffield, P.J, Aoki, J, Martin, T.W, Arai, H, Inoue, K, Derewenda, Z.S. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib).
Protein Eng., 13, 2000
|
|
1E0W
 
 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1E0X
 
 | | XYLANASE 10A FROM SREPTOMYCES LIVIDANS. XYLOBIOSYL-ENZYME INTERMEDIATE AT 1.65 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1E0V
 
 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
3DT9
 
 | | Crystal Structure of Bovin Brain Platelet Activating Factor Acetylhydrolase Covalently Inhibited by Soman | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (R)-METHYLPHOSPHINATE, Brain Platelet-activating factor acetylhydrolase IB subunit alpha | | Authors: | Epstein, T.M, Samanta, U, Bahnson, B.J. | | Deposit date: | 2008-07-14 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of brain group-VIII phospholipase A2 in nonaged complexes with the organophosphorus nerve agents soman and sarin.
Biochemistry, 48, 2009
|
|
3DT8
 
 | |
3DT6
 
 | |
4YTF
 
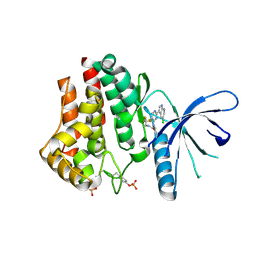 | | Discovery of VX-509 (Decernotinib): A Potent and Selective Janus kinase (JAK) 3 Inhibitor for the Treatment of Autoimmune Diseases | | Descriptor: | N~2~-[2-(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-fluoropyrimidin-4-yl]-N-(2,2,2-trifluoroethyl)-L-alaninamide, Tyrosine-protein kinase JAK2 | | Authors: | Farmer, L, Ledeboer, M.W, Zuccola, H.J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of VX-509 (Decernotinib): A Potent and Selective Janus Kinase 3 Inhibitor for the Treatment of Autoimmune Diseases.
J.Med.Chem., 58, 2015
|
|
4YTI
 
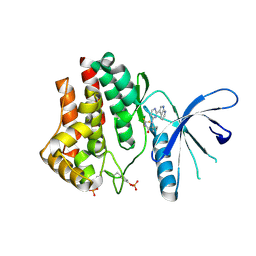 | |
4YTC
 
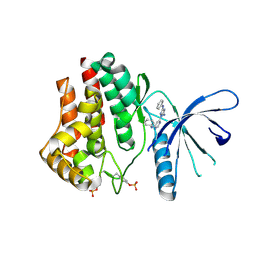 | |
4YTH
 
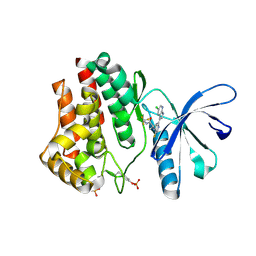 | |
1C8U
 
 | |
2JRT
 
 | | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides. Northeast Structural Genomics target RhR5 | | Descriptor: | Uncharacterized protein | | Authors: | Wang, L, Chen, C, Nwosu, C, Cunningham, K, Owens, L, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides.
To be Published
|
|
3KA7
 
 | | Crystal Structure of an oxidoreductase from Methanosarcina mazei. Northeast Structural Genomics Consortium target id MaR208 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-18 | | Release date: | 2009-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an oxidoreductase from Methanosarcina mazei
To be Published
|
|
2QS9
 
 | | Crystal structure of the human retinoblastoma-binding protein 9 (RBBP-9). NESG target HR2978 | | Descriptor: | Retinoblastoma-binding protein 9 | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Kuzin, A, Chen, C.X, Cunningham, K, Owens, L, Maglaqui, M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of human retinoblastoma binding protein 9.
Proteins, 74, 2008
|
|
2ESM
 
 | | Crystal Structure of ROCK 1 bound to fasudil | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, Rho-associated protein kinase 1 | | Authors: | Jacobs, M. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity.
J.Biol.Chem., 281, 2006
|
|
2ETK
 
 | | Crystal Structure of ROCK 1 bound to hydroxyfasudil | | Descriptor: | 1-(1-HYDROXY-5-ISOQUINOLINESULFONYL)HOMOPIPERAZINE, Rho-associated protein kinase 1 | | Authors: | Jacobs, M. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity.
J.Biol.Chem., 281, 2006
|
|
2ETR
 
 | | Crystal Structure of ROCK I bound to Y-27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 1 | | Authors: | Jacobs, M. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity.
J.Biol.Chem., 281, 2006
|
|
2ERZ
 
 | | Crystal Structure of c-AMP Dependent Kinase (PKA) bound to hydroxyfasudil | | Descriptor: | 1-(1-HYDROXY-5-ISOQUINOLINESULFONYL)HOMOPIPERAZINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Jacobs, M. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity.
J.Biol.Chem., 281, 2006
|
|
