8VAO
 
 | |
8VCR
 
 | | SARS-CoV-2 Spike S2 bound to Fab 54043-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 54043-5 Fab Heavy Chain, ... | | Authors: | Johnson, N.V, McLellan, J.S. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Discovery and characterization of a pan-betacoronavirus S2-binding antibody.
Structure, 2024
|
|
8V5A
 
 | |
5EA8
 
 | | Crystal Structure of Prefusion RSV F Glycoprotein Fusion Inhibitor Resistance Mutant D489Y | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, D(-)-TARTARIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
4ZYP
 
 | | Crystal Structure of Motavizumab and Quaternary-Specific RSV-Neutralizing Human Antibody AM14 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | AM14 antibody Fab heavy chain, AM14 antibody light chain, Fusion glycoprotein F0,Fibritin, ... | | Authors: | Gilman, M.S.A, McLellan, J.S. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein.
Plos Pathog., 11, 2015
|
|
5EA4
 
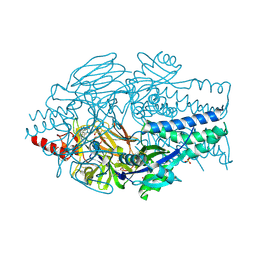 | | Crystal Structure of Inhibitor JNJ-49153390 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3-[[5-bromanyl-1-(3-methylsulfonylpropyl)benzimidazol-2-yl]methyl]-1-cyclopropyl-imidazo[4,5-c]pyridin-2-one, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5EA3
 
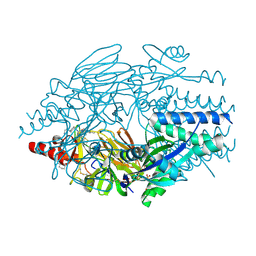 | | Crystal Structure of Inhibitor JNJ-2408068 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-[[2-[[1-(2-azanylethyl)piperidin-4-yl]amino]-4-methyl-benzimidazol-1-yl]methyl]-6-methyl-pyridin-3-ol, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Langedijk, J.P, Roymans, D. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5FHC
 
 | |
5FHB
 
 | |
5EA6
 
 | | Crystal Structure of Inhibitor BTA-9881 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | (9~{b}~{R})-9~{b}-(4-chlorophenyl)-1-pyridin-3-ylcarbonyl-2,3-dihydroimidazo[5,6]pyrrolo[1,2-~{a}]pyridin-5-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5EA5
 
 | | Crystal Structure of Inhibitor TMC-353121 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-[[6-[[[2-(3-hydroxypropyl)-5-methylphenyl]amino]methyl]-2-[[3-(4-morpholinyl)propyl]amino]-1H-benzimidazol-1-yl]methyl]-6-methyl-3-pyridinol, D(-)-TARTARIC ACID, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5FHA
 
 | |
5EA7
 
 | | Crystal Structure of Inhibitor BMS-433771 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1-cyclopropyl-3-[[1-(4-oxidanylbutyl)benzimidazol-2-yl]methyl]imidazo[4,5-c]pyridin-2-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
6BLI
 
 | | RSV G peptide bound to Fab CB002.5 | | Descriptor: | CB002.5 Fab Heavy Chain, CB002.5 Fab Light Chain, Major surface glycoprotein G | | Authors: | Jones, H.G, McLellan, J.S, Langedijk, J.P. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies.
PLoS Pathog., 14, 2018
|
|
6BLH
 
 | | RSV G central conserved region bound to Fab CB017.5 | | Descriptor: | 1,2-ETHANEDIOL, Fab CB017.5 heavy chain, Fab CB017.5 light chain, ... | | Authors: | Jones, H.G, McLellan, J.S, Langedijk, J.P. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies.
PLoS Pathog., 14, 2018
|
|
7T01
 
 | | SARS-CoV-2 S-RBD + Fab 54042-4 | | Descriptor: | 54042-4 Fab - Heavy Chain, 54042-4 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Potent neutralization of SARS-CoV-2 variants of concern by an antibody with an uncommon genetic signature and structural mode of spike recognition.
Cell Rep, 37, 2021
|
|
7T67
 
 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with One(1) RBD Up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
7T3M
 
 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, composite map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2-7 scFv, ... | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
6U7K
 
 | | Prefusion structure of PEDV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-09-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The 3.1-Angstrom Cryo-electron Microscopy Structure of the Porcine Epidemic Diarrhea Virus Spike Protein in the Prefusion Conformation.
J.Virol., 93, 2019
|
|
8TEA
 
 | |
6UOE
 
 | |
7USL
 
 | | Integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin.
Cell Rep, 40, 2022
|
|
7USM
 
 | | Integrin alphaM/beta2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin.
Cell Rep, 40, 2022
|
|
7SSC
 
 | |
7TJQ
 
 | | SAN27-14 bound to a antigenic site V on prefusion-stabilized hMPV F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, MPE8 Fab heavy chain, ... | | Authors: | Hsieh, C.-L, McLellan, J.S, Rush, S.A. | | Deposit date: | 2022-01-16 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Characterization of prefusion-F-specific antibodies elicited by natural infection with human metapneumovirus.
Cell Rep, 40, 2022
|
|
