5FOG
 
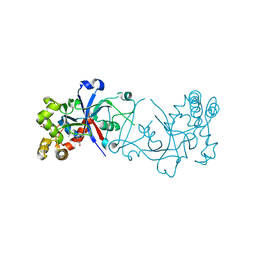 | | Crystal structure of hte Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of norvaline (Nv2AA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOL
 
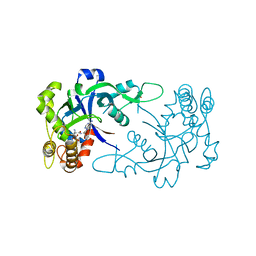 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of isoeucine (Ile2AA) | | Descriptor: | 2'-(L-ISOLEUCYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, PHOSPHATE ION | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FON
 
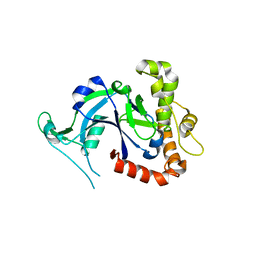 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain (apo structure) | | Descriptor: | LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
8UZH
 
 | |
8UQV
 
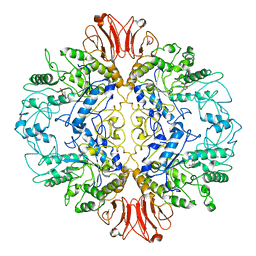 | |
6TPR
 
 | | PqsR (MvfR) bound to inhibitory compound 40 | | Descriptor: | 2-[(5-methyl-[1,2,4]triazino[5,6-b]indol-3-yl)sulfanyl]-~{N}-(4-pyridin-2-yloxyphenyl)ethanamide, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hit Identification of New Potent PqsR Antagonists as Inhibitors of Quorum Sensing in Planktonic and Biofilm GrownPseudomonas aeruginosa.
Front Chem, 8, 2020
|
|
7D3Y
 
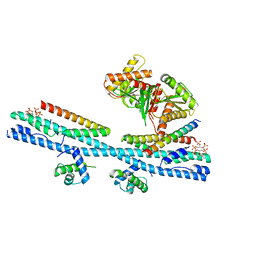 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
7D3T
 
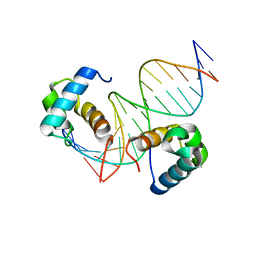 | | Crystal structure of OSPHR2 in complex with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*CP*GP*GP*AP*TP*AP*TP*CP*CP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*GP*CP*TP*TP*GP*AP*GP*GP*AP*TP*AP*TP*CP*CP*GP*A)-3'), Protein PHOSPHATE STARVATION RESPONSE 2 | | Authors: | Guan, Z.Y, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
5CI9
 
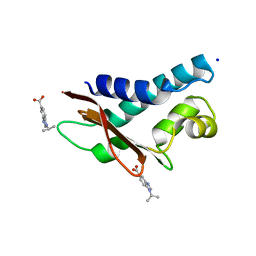 | | Crystal structure of human Tob in complex with inhibitor fragment 6 | | Descriptor: | 1-(propan-2-yl)-1H-benzimidazole-5-carboxylic acid, Protein Tob1, SODIUM ION | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
7CVQ
 
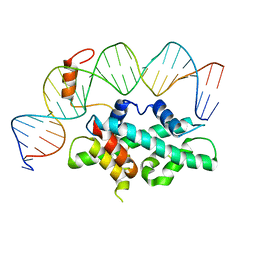 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
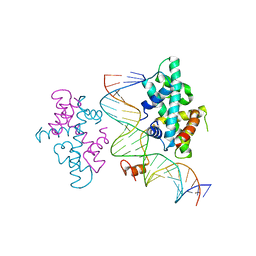 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
5CI8
 
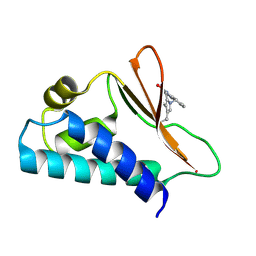 | | Crystal structure of human Tob in complex with inhibitor fragment 1 | | Descriptor: | Protein Tob1, pyrrolo[1,2-a]quinoxalin-4(5H)-one | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2015-12-09 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
4ZHU
 
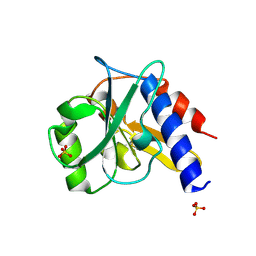 | | Crystal structure of a bacterial repressor protein | | Descriptor: | SULFATE ION, YfiR | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.3968 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4ZHW
 
 | |
4ZHY
 
 | | Crystal structure of a bacterial signalling complex | | Descriptor: | FORMIC ACID, SULFATE ION, YfiB, ... | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4WY4
 
 | | Crystal structure of autophagic SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Zhao, M, Brunger, A.T. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes.
Nature, 520, 2015
|
|
4Z3D
 
 | |
4ZHV
 
 | | Crystal structure of a bacterial signalling protein | | Descriptor: | SULFATE ION, YfiB | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
7X78
 
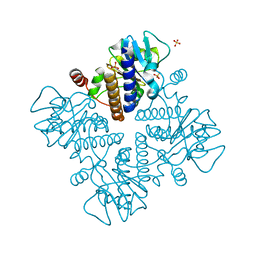 | | L-fuculose 1-phosphate aldolase | | Descriptor: | L-fuculose phosphate aldolase, MAGNESIUM ION, SULFATE ION | | Authors: | Lou, X, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of an L-fuculose-1-phosphate aldolase from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 607, 2022
|
|
5DJU
 
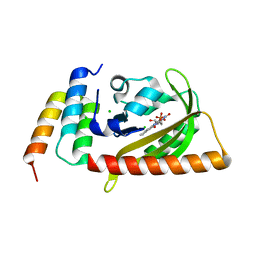 | | Crystal structure of LOV2 (C450A) domain in complex with Zdk3 | | Descriptor: | CHLORIDE ION, Engineered protein, Zdk3 affibody, ... | | Authors: | Tarnawski, M, Wang, H, Yumerefendi, H, Hahn, K.M, Schlichting, I. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
5DV2
 
 | |
7YTJ
 
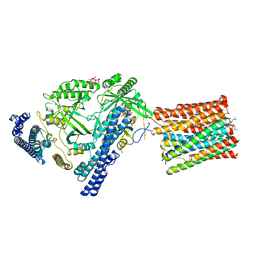 | | Cryo-EM structure of VTC complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
3F38
 
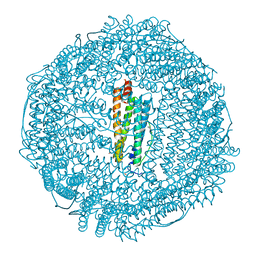 | | Apoferritin: complex with 2,6-dimethylphenol | | Descriptor: | 2,6-dimethylphenol, CADMIUM ION, Ferritin light chain, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
