3RMU
 
 | | Crystal structure of human Methylmalonyl-CoA epimerase, MCEE | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Methylmalonyl-CoA epimerase, ... | | Authors: | Chaikuad, A, Krysztofinska, E, Froese, D.S, Yue, W.W, Vollmar, M, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Methylmalonyl-CoA epimerase, MCEE
To be Published
|
|
3PEJ
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of Macbecin | | Descriptor: | Endoplasmin homolog, MACBECIN, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Hutchinson, A, Weadge, J, MacKenzie, F, Senisterra, G, Vedadi, M, Cossar, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Pizzaro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of Macbecin
To be Published
|
|
3PEH
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative | | Descriptor: | 2-amino-4-{2,4-dichloro-5-[2-(diethylamino)ethoxy]phenyl}-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, Endoplasmin homolog, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Hutchinson, A, Weadge, J, Cossar, D, MacKenzie, F, Vedadi, M, Senisterra, G, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative
To be Published
|
|
3PFY
 
 | | The catalytic domain of human OTUD5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, OTU domain-containing protein 5, SULFATE ION, ... | | Authors: | Walker, J.R, Asinas, A.E, Crombet, L, Dong, A, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | The catalytic domain of human OTUD5
To be Published
|
|
3PFV
 
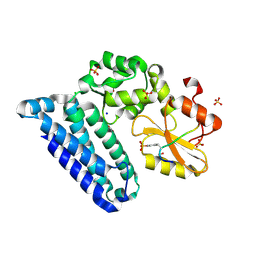 | | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide | | Descriptor: | 1,2-ETHANEDIOL, 11-meric peptide from Epidermal growth factor receptor, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Guo, K, Cooper, C.D.O, Ayinampudi, V, Krojer, T, Muniz, J.R.C, Vollmar, M, Canning, P, Gileadi, O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide
To be Published
|
|
3PFN
 
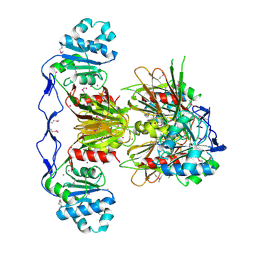 | | Crystal Structure of human NAD kinase | | Descriptor: | NAD kinase, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Tempel, W, Wernimont, A.K, Tong, Y, Guan, X, Shen, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of human NAD kinase
to be published
|
|
3OLH
 
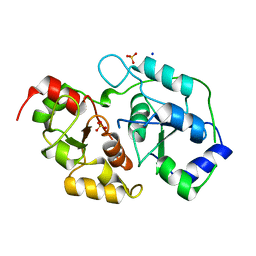 | | Human 3-mercaptopyruvate sulfurtransferase | | Descriptor: | 3-mercaptopyruvate sulfurtransferase, SODIUM ION, SULFATE ION | | Authors: | Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human 3-Mercaptopyruvate Sulfurtransferase
To be Published
|
|
3OOY
 
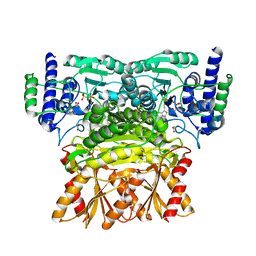 | | Crystal structure of human Transketolase (TKT) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Krojer, T, Krysztofinska, E, Guo, K, Pilka, E, Kochan, G, Chaikuad, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Transketolase (TKT)
To be Published
|
|
3OOM
 
 | | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507 | | Descriptor: | 1,2-ETHANEDIOL, 1-{3-[6-(tetrahydro-2H-pyran-4-ylamino)imidazo[1,2-b]pyridazin-3-yl]phenyl}ethanone, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C, Krojer, T, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507
To be Published
|
|
3OZ6
 
 | | Crystal structure of MapK from Cryptosporidium Parvum, cgd2_1960 | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, serine/threonine protein kinase | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Sullivan, H, Hassanali, A, Kozieradzki, I, Cossar, D, Arrowsmith, C.H, Bochkarev, A, Bountra, C, Edwards, A.M, Weigelt, J, Hui, R, Neculai, A.M, Hutchinson, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of MapK from Cryptosporidium Parvum, cgd2_1960
To be Published
|
|
3S8S
 
 | | Crystal structure of the RRM domain of human SETD1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, UNKNOWN ATOM OR ION | | Authors: | Chao, X, Tempel, W, Bian, C, Cerovina, T, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-30 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the RRM domain of human SETD1A
to be published
|
|
5DVR
 
 | | Crystal Structure of TgCDPK1 From Toxoplasma Gondii complexed with GW780159X | | Descriptor: | 4-(3-chlorophenyl)-5-(1,5-naphthyridin-2-yl)-1,3-thiazol-2-amine, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lovato, D.V, Osman, K.T, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-21 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of TgCDPK1 From Toxoplasma Gondii complexed with GW780159X
to be published
|
|
5E8R
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | CHLORIDE ION, N-methyl-N-({4-[4-(propan-2-yloxy)phenyl]-1H-pyrrol-3-yl}methyl)ethane-1,2-diamine, Protein arginine N-methyltransferase 6, ... | | Authors: | DONG, A, ZENG, H, LIU, J, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, JIN, J, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases.
Acs Chem.Biol., 11, 2016
|
|
5DKC
 
 | | Crystal structure of the bromodomain of human BRM (SMARCA2) in complex with PFI-3 chemical probe | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(1R,4R)-5-(pyridin-2-yl)-2,5-diazabicyclo[2.2.1]hept-2-yl]prop-2-en-1-one, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Fedorov, O, Savitsky, P, Nunez-Alonso, G, Fonseca, M, Krojer, T, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the bromodomain of human BRM (SMARCA2) in complex with PFI-3 chemical probe
To Be Published
|
|
5E9V
 
 | | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(imidazo[1,2-a]pyridin-5-yl)-7-(morpholin-4-yl)indolizin-3-yl]ethanone, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Hay, D.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Schofield, C.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand
To Be Published
|
|
5EAL
 
 | | Crystal structure of human WDR5 in complex with compound 9h | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-quinolin-3-yl-phenyl]benzamide, CHLORIDE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human WDR5 in complex with compound 9h
to be published
|
|
5E26
 
 | | Crystal structure of human PANK2: the catalytic core domain in complex with pantothenate and adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, LOPPNAU, P, RAVICHANDRAN, M, CHENG, C, TEMPEL, W, SEITOVA, A, HUTCHINSON, A, HONG, B.S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of human PANK2: the catalytic core domain in complex with pantothenate and adenosine diphosphate
to be published
|
|
5EPL
 
 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EZR
 
 | | Crystal Structure of PVX_084705 bound to compound | | Descriptor: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | Authors: | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
5F0A
 
 | | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine INHIBITOR | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, cGMP-dependent protein kinase, putative | | Authors: | Walker, J.R, Wernimont, A.K, He, H, Seitova, A, Loppnau, P, Sibley, L.D, Graslund, S, Hutchinson, A, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Hui, R, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND INHIBITOR
To be published
|
|
5FYB
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with MC1648 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Rotili, D, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-05 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Mc1648
To be Published
|
|
5FZD
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with L-2-hydroxyglutarate | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with L-2-Hydroxyglutarate
To be Published
|
|
5FAI
 
 | | EMG1 N1-Specific Pseudouridine Methyltransferase | | Descriptor: | CITRIC ACID, Ribosomal RNA small subunit methyltransferase NEP1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | DONG, A, ZENG, H, LI, Y, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | EMG1 N1-Specific Pseudouridine Methyltransferase
to be published
|
|
5FYM
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with D-2-hydroxyglutarate | | Descriptor: | (2R)-2-hydroxypentanedioic acid, 1,2-ETHANEDIOL, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with D-2- Hydroxyglutarate
To be Published
|
|
5FZ7
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment ethyl 2-amino-4-thiophen-2-ylthiophene-3- carboxylate (N06131b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Ethyl 2-Amino-4-Thiophen-2-Ylthiophene-3-Carboxylate (N06131B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
