7EPA
 
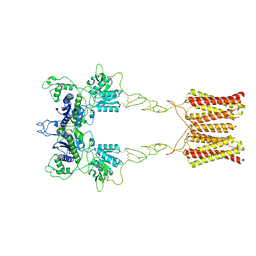 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPB
 
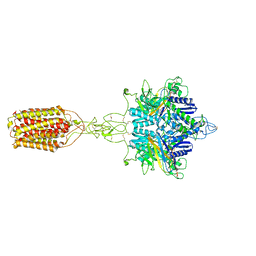 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPC
 
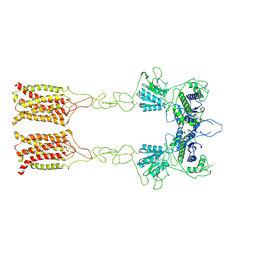 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | Descriptor: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
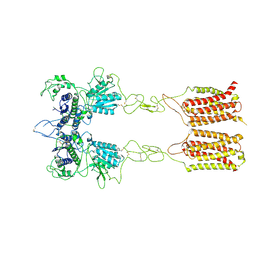 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPF
 
 | | Crystal structure of mGlu2 bound to NAM597 | | Descriptor: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
2QN1
 
 | | Glycogen Phosphorylase b in complex with asiatic acid | | Descriptor: | Glycogen phosphorylase, muscle form, asiatic acid | | Authors: | Zographos, S.E, Leonidas, D.D, Alexacou, K.-M, Hayes, J, Oikonomakos, N.G. | | Deposit date: | 2007-07-17 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Naturally occurring pentacyclic triterpenes as inhibitors of glycogen phosphorylase: synthesis, structure-activity relationships, and X-ray crystallographic studies
J.Med.Chem., 51, 2008
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
5X04
 
 | | 12:0-ACP thioesterase from Umbellularia californica | | Descriptor: | Dodecanoyl-[acyl-carrier-protein] hydrolase, chloroplastic | | Authors: | Xue, S, Feng, Y. | | Deposit date: | 2017-01-19 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insight into Acyl-ACP Thioesterase toward Substrate Specificity Design.
ACS Chem. Biol., 12, 2017
|
|
4OD0
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
2QN2
 
 | | Glycogen Phosphorylase b in complex with Maslinic Acid | | Descriptor: | Glycogen phosphorylase, muscle form, maslinic acid | | Authors: | Zographos, S.E, Leonidas, D.D, Alexacou, K.-M, Hayes, J, Oikonomakos, N.G. | | Deposit date: | 2007-07-17 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Naturally occurring pentacyclic triterpenes as inhibitors of glycogen phosphorylase: synthesis, structure-activity relationships, and X-ray crystallographic studies
J.Med.Chem., 51, 2008
|
|
2B53
 
 | |
2NWT
 
 | | NMR Structure of Protein UPF0165 protein AF_2212 from Archaeoglobus Fulgidus; Northeast Structural Genomics Consortium Target GR83 | | Descriptor: | UPF0165 protein AF_2212 | | Authors: | Singarapu, K.K, Sukumaran, D.K, Parish, D, Atreya, H.S, Liu, G, Eletsky, A, Chen, C.X, Jiang, M, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Protein Y2212_ARCFU from Archaeoglobus Fulgidus; Northeast Structural Genomics Consortium Target GR83
To be Published
|
|
8EHI
 
 | | Cryo-EM structure of his-elemental paused elongation complex with an unfolded TL (2) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EHF
 
 | | Cryo-EM structure of his-elemental paused elongation complex with an unfolded TL (1) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EG8
 
 | | Cryo-EM structure of consensus elemental paused elongation complex with a folded TL | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EHA
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (out) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EGB
 
 | | Cryo-EM structure of consensus elemental paused elongation complex with an unfolded TL | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EG7
 
 | | Cryo-EM structure of pre-consensus elemental paused elongation complex | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-11 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EH8
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (1) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EH9
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (2) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WKJ
 
 | | A COVID-19 T-cell response detection method based on a newly identified human CD8+ T cell epitope from SARS-CoV-2-Hubei Province, 2021. | | Descriptor: | Beta-2-microglobulin, LYS-THR-PHE-PRO-PRO-THR-GLU-PRO-LYS, MHC class I antigen | | Authors: | Zhang, J, Lu, D, Li, M, Liu, M.S, Yao, S.J, Zhan, J.B, Liu, J, Gao, G.F. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
China CDC Wkly, 4, 2022
|
|
7TXN
 
 | | Reduced Structure of RexT | | Descriptor: | CHLORIDE ION, GLYCEROL, Transcriptional regulator | | Authors: | Bridwell-Rabb, J, Li, B. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT.
Commun Biol, 5, 2022
|
|
7TXM
 
 | | Oxidized Structure of RexT | | Descriptor: | HYDROGEN PEROXIDE, Transcriptional regulator | | Authors: | Bridwell-Rabb, J, Li, B. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT.
Commun Biol, 5, 2022
|
|
7TXO
 
 | | Selenomethionine Labeled Structure of RexT | | Descriptor: | CHLORIDE ION, Transcriptional regulator | | Authors: | Bridwell-Rabb, J, Li, B. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT.
Commun Biol, 5, 2022
|
|
