2QR5
 
 | | Aeropyrum pernix acylaminoacyl peptidase, H367A mutant | | Descriptor: | Acylamino-acid-releasing enzyme | | Authors: | Harmat, V, Pallo, A, Kiss, A.L, Polgar, L. | | Deposit date: | 2007-07-27 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and kinetic contributions of the oxyanion binding site to the catalytic activity of acylaminoacyl peptidase
J.Struct.Biol., 162, 2008
|
|
2QY0
 
 | | Active dimeric structure of the catalytic domain of C1r reveals enzyme-product like contacts | | Descriptor: | Complement C1r subcomponent, GLYCEROL | | Authors: | Kardos, J, Harmat, V, Pallo, A, Barabas, O, Szilagyi, K, Graf, L, Naray-Szabo, G, Goto, Y, Zavodszky, P, Gal, P. | | Deposit date: | 2007-08-13 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r.
Mol.Immunol., 45, 2008
|
|
8W5B
 
 | | Crystal Structure of the shaft pilin LrpA from Ligilactobacillus ruminis | | Descriptor: | IODIDE ION, LPXTG-motif cell wall anchor domain protein | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WB8
 
 | |
4UX9
 
 | | Crystal structure of JNK1 bound to a MKK7 docking motif | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7, MITOGEN-ACTIVATED PROTEIN KINASE 8, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kragelj, J, Palencia, A, Nanao, M.H, Maurin, D, Bouvignies, G, Blackledge, M, Ringkjobing-Jensen, M. | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Dynamics of the Mkk7-Jnk Signaling Complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6M48
 
 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - P21212 form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SpaC | | Authors: | Kant, A, Palva, A, Von Ossowaski, I, Krishnan, V. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
6M3Y
 
 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - open conformation | | Descriptor: | MAGNESIUM ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
6M7C
 
 | |
4V2B
 
 | | rat Unc5D Ig domain 1 | | Descriptor: | PROTEIN UNC5D | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
7BVX
 
 | | Crystal structure of C-terminal fragment of pilus adhesin SpaC from Lactobacillus rhamnosus GG-Iodide soaked | | Descriptor: | IODIDE ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-04-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
4AFL
 
 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
8KB2
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - iodide derivative | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8KG4
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - orthorhombic form | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-17 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8KCL
 
 | |
1OLL
 
 | | Extracellular region of the human receptor NKp46 | | Descriptor: | 1,2-ETHANEDIOL, NK RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Pesce, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-08-07 | | Release date: | 2003-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the Human Nk Cell Triggering Receptor Nkp46 Ectodomain
Biochem.Biophys.Res.Commun., 309, 2003
|
|
3ZGZ
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and toxic moiety from agrocin 84 (TM84) in aminoacylation-like conformation | | Descriptor: | LEUCINE--TRNA LIGASE, MAGNESIUM ION, TRNA-LEU UAA ISOACCEPTOR, ... | | Authors: | Chopra, S, Palencia, A, Virus, C, Tripathy, A, Temple, B.R, Velazquez-Campoy, A, Cusack, S, Reader, J.S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plant Tumour Biocontrol Agent Employs a tRNA-Dependent Mechanism to Inhibit Leucyl-tRNA Synthetase
Nat.Commun., 4, 2013
|
|
3ZJV
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJT
 
 | | Ternary complex of E.coli leucyl-tRNA synthetase, tRNA(Leu)574 and the benzoxaborole AN3017 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJU
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3016 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
6LPF
 
 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6LR6
 
 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
1HKF
 
 | | The three dimensional structure of NK cell receptor Nkp44, a triggering partner in natural cytotoxicity | | Descriptor: | NK CELL ACTIVATING RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-03-10 | | Release date: | 2003-06-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Three-Dimensional Structure of the Human Nk Cell Receptor Nkp44, a Triggering Partner in Natural Cytotoxicity
Structure, 11, 2003
|
|
1H4K
 
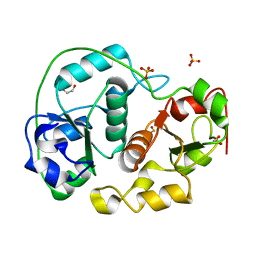 | | Sulfurtransferase from Azotobacter vinelandii in complex with hypophosphite | | Descriptor: | 1,2-ETHANEDIOL, HYPOPHOSPHITE, SULFATE ION, ... | | Authors: | Bordo, D, Forlani, F, Spallarossa, A, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Persulfurated Cysteine Promotes Active Site Reactivity in Azotobacter Vinelandii Rhodanse
Biol.Chem., 382, 2001
|
|
1H4M
 
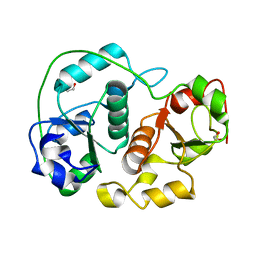 | | Sulfurtransferase from Azotobacter vinelandii in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PUTATIVE THIOSULFATE SULFURTRANSFERASE | | Authors: | Bordo, D, Forlani, F, Spallarossa, A, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Persulfurated Cysteine Promotes Active Site Reactivity in Azotobacter Vinelandii Rhodanse
Biol.Chem., 382, 2001
|
|
4B9W
 
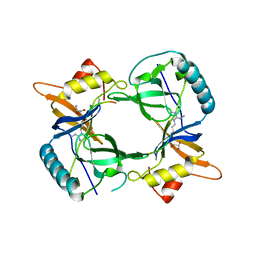 | | Structure of extended Tudor domain TD3 from mouse TDRD1 in complex with MILI peptide containing dimethylarginine 45. | | Descriptor: | GLYCEROL, PIWI-LIKE PROTEIN 2, TUDOR DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | Deposit date: | 2012-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors.
Rna, 18, 2012
|
|
