2H4V
 
 | | Crystal Structure of the Human Tyrosine Receptor Phosphatase Gamma | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ugochukwu, E, Barr, A, Das, S, Eswaran, J, Savitsky, P, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Debreczeni, J, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2GW2
 
 | | Crystal structure of the peptidyl-prolyl isomerase domain of human cyclophilin G | | Descriptor: | Peptidyl-prolyl cis-trans isomerase G, UNKNOWN ATOM OR ION | | Authors: | Bernstein, G, Tempel, W, Davis, T, Newman, E.M, Finerty Jr, P.J, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2NLK
 
 | | Crystal structure of D1 and D2 catalytic domains of human Protein Tyrosine Phosphatase Gamma (D1+D2 PTPRG) | | Descriptor: | Protein tyrosine phosphatase, receptor type, G variant (Fragment) | | Authors: | Filippakopoulos, P, Gileadi, O, Johansson, C, Ugochukwu, E, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-20 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2NPM
 
 | | crystal structure of Cryptosporidium parvum 14-3-3 protein in complex with peptide | | Descriptor: | 14-3-3 domain containing protein, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Dong, A, Lew, J, Wasney, G, Ren, H, Lin, L, Hassanali, A, Qiu, W, Zhao, Y, Doyle, D, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
2HE9
 
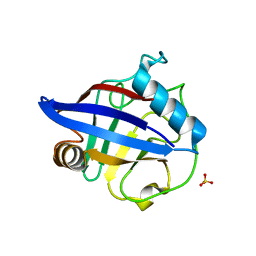 | | Structure of the peptidylprolyl isomerase domain of the human NK-tumour recognition protein | | Descriptor: | NK-tumor recognition protein, SULFATE ION | | Authors: | Walker, J.R, Davis, T, Newman, E.M, MacKenzie, F, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2HQ6
 
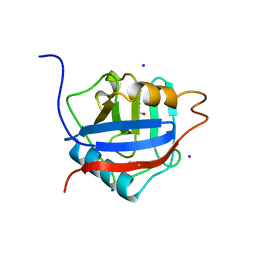 | | Structure of the Cyclophilin_CeCYP16-Like Domain of the Serologically Defined Colon Cancer Antigen 10 from Homo Sapiens | | Descriptor: | GLYCEROL, IODIDE ION, Serologically defined colon cancer antigen 10 | | Authors: | Walker, J.R, Davis, T, Paramanathan, R, Newman, E.M, Finerty Jr, P.J, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2NZ6
 
 | | Crystal structure of the PTPRJ inactivating mutant C1239S | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Ugochukwu, E, Barr, A, Savitsky, P, Pike, A.C.W, Bunkoczi, G, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2OC3
 
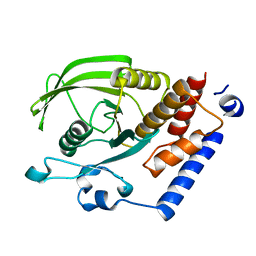 | | Crystal Structure of the Catalytic Domain of Human Protein Tyrosine Phosphatase non-receptor Type 18 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Ugochukwu, E, Barr, A, Alfano, I, Gorrec, F, Umeano, C, Savitsky, P, Sobott, F, Eswaran, J, Papagrigoriou, E, Debreczeni, J.E, Turnbull, A, Bunkoczi, G, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2I75
 
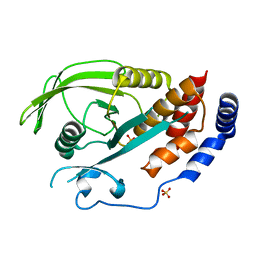 | | Crystal Structure of Human Protein Tyrosine Phosphatase N4 (PTPN4) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Ugochukwu, E, Barr, A, Savitsky, P, Burgess, N, Das, S, Turnbull, A, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
8E1P
 
 | | Crystal structure of BG505 SOSIP.v4.1-GT1.2 trimer in complex with gl-PGV20 and PGT124 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505-SOSIP.v4.1-GT1.2gp120, ... | | Authors: | Sarkar, A, Kumar, S, Wilson, I.A. | | Deposit date: | 2022-08-11 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.82 Å) | | Cite: | Germline-targeting HIV-1 Env vaccination induces VRC01-class antibodies with rare insertions.
Cell Rep Med, 4, 2023
|
|
8DYT
 
 | | Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide | | Descriptor: | 227 Fab heavy chain, 227 Fab light chain, Circumsporozoite protein | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Affinity-matured homotypic interactions induce spectrum of PfCSP structures that influence protection from malaria infection.
Nat Commun, 14, 2023
|
|
8DYW
 
 | |
8DYY
 
 | |
8DZ3
 
 | |
8DZ5
 
 | |
8DYX
 
 | |
8DZ4
 
 | |
8EH5
 
 | | Cryo-EM structure of L9 Fab in complex with rsCSP | | Descriptor: | Circumsporozoite protein, L9 Heavy chain, L9 Light chain | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-09-13 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of epitope selectivity and potent protection from malaria by PfCSP antibody L9.
Nat Commun, 14, 2023
|
|
8EKF
 
 | |
4KSA
 
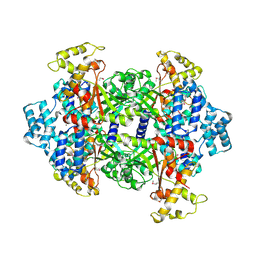 | | Crystal Structure of Malonyl-CoA decarboxylase from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR127 | | Descriptor: | MAGNESIUM ION, Malonyl-CoA decarboxylase | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
4KS9
 
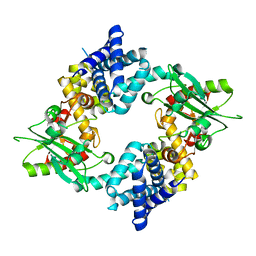 | | Crystal Structure of Malonyl-CoA decarboxylase (Rmet_2797) from Cupriavidus metallidurans, Northeast Structural Genomics Consortium Target CrR76 | | Descriptor: | MAGNESIUM ION, Malonyl-CoA decarboxylase | | Authors: | Forouhar, F, Tran, T.H, Lew, S, Seetharaman, J, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
1ZKC
 
 | | Crystal Structure of the cyclophiln_RING domain of human peptidylprolyl isomerase (cyclophilin)-like 2 isoform b | | Descriptor: | BETA-MERCAPTOETHANOL, Peptidyl-prolyl cis-trans isomerase like 2 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2A8B
 
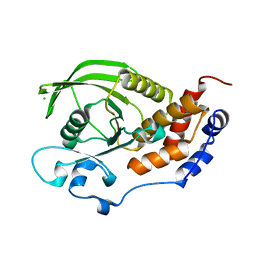 | | Crystal Structure of the Catalytic Domain of Human Tyrosine Phosphatase Receptor, Type R | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase R | | Authors: | Ugochukwu, E, Eswaran, J, Barr, A, Longman, E, Arrowsmith, C, Edwards, A, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-07 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases.
Biochem.J., 395, 2006
|
|
2B49
 
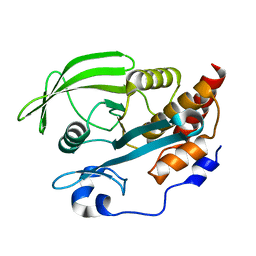 | | Crystal Structure of the Catalytic Domain of Protein Tyrosine Phosphatase, non-receptor Type 3 | | Descriptor: | protein tyrosine phosphatase, non-receptor type 3 | | Authors: | Ugochukwu, E, Arrowsmith, C, Barr, A, Bunkoczi, G, Das, S, Debreczeni, J, Edwards, A, Eswaran, J, Knapp, S, Sundstrom, M, Turnbull, A, von Delft, F, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2BTP
 
 | | 14-3-3 Protein Theta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN TAU, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Johansson, A.C.E, Smee, C, Yang, X, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-05 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
