6O68
 
 | |
6ONI
 
 | |
6PDZ
 
 | |
7JHG
 
 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JHH
 
 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Fab and nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JIJ
 
 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Powell, K, Xu, T, Brunzelle, J.S, Xu, H.X, Melcher, K. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
4IS8
 
 | |
5SYZ
 
 | | Human Liver Receptor Homologue-1 (LRH-1) Bound to a RJW100 stereoisomer and a Fragment of TIF-2 | | Descriptor: | (1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Ortlund, E.A, Mays, S.G. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9261 Å) | | Cite: | Crystal Structures of the Nuclear Receptor, Liver Receptor Homolog 1, Bound to Synthetic Agonists.
J. Biol. Chem., 291, 2016
|
|
5TO5
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO6
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Bandyopadhyay, A, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO7
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TVB
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
4K4J
 
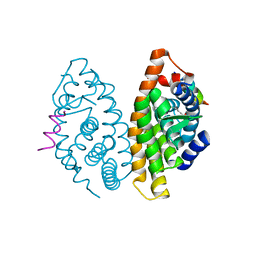 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 9cUAB30 and the coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-8-(3,4-dihydronaphthalen-1(2H)-ylidene)-3,7-dimethylocta-2,4,6-trienoic acid, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Defining the Communication between Agonist and Coactivator Binding in the Retinoid X Receptor alpha Ligand Binding Domain.
J.Biol.Chem., 289, 2014
|
|
4K6I
 
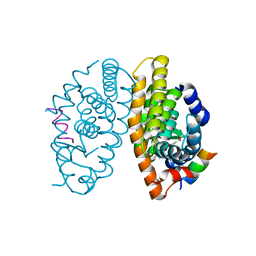 | | Crystal structure of human retinoid X receptor alpha-ligand binding domain complex with Targretin and the coactivator peptide GRIP-1 | | Descriptor: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2013-04-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining the Communication between Agonist and Coactivator Binding in the Retinoid X Receptor alpha Ligand Binding Domain.
J.Biol.Chem., 289, 2014
|
|
5UNJ
 
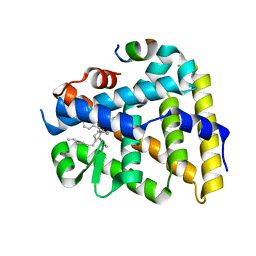 | | Structure of Human Liver Receptor Homolog 1 in complex with PGC1a and RJW100 | | Descriptor: | (1R,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor subfamily 5 group A member 2, Peroxisome proliferator-activated gamma coactivator 1-alpha | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Dynamics of the Liver Receptor Homolog 1-PGC1 alpha Complex.
Mol. Pharmacol., 92, 2017
|
|
5V39
 
 | |
4LKP
 
 | |
4LKT
 
 | |
4YWC
 
 | | Crystal structure of Myc3(5-242) fragment in complex with Jaz9(218-239) peptide | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2015-03-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
7SNQ
 
 | |
7SQB
 
 | | PPAR gamma LBD bound to Inverse Agonist SR10221 | | Descriptor: | (2S)-2-{5-[(5-{[(1S)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-11-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PPAR gamma Corepression Involves Alternate Ligand Conformation and Inflation of H12 Ensembles.
Acs Chem.Biol., 18, 2023
|
|
7SQA
 
 | | PPAR gamma LBD bound to SR10221 and SMRT corepressor motif | | Descriptor: | (2S)-2-{5-[(5-{[(1S)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-11-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | PPAR gamma Corepression Involves Alternate Ligand Conformation and Inflation of H12 Ensembles.
Acs Chem.Biol., 18, 2023
|
|
7TT8
 
 | | Human LRH-1 LBD bound to agonist 6N-10CA and fragment of Tif2 coactivator | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-01-31 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Modulation of Nuclear Receptor LRH-1 through Targeting Buried and Surface Regions of the Binding Pocket.
J.Med.Chem., 65, 2022
|
|
4YZ6
 
 | | Crystal Structure of Myc3[44-238] from Arabidopsis in complex with Jaz1 peptide [200-221] | | Descriptor: | Protein TIFY 10A, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J, Xu, H.E, Melcher, K, HE, S.Y. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
8CVP
 
 | | Cereblon-DDB1 in the Apo form | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, ZINC ION | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
