6UXR
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with LysoPC | | Descriptor: | Bcl-2 homologous antagonist/killer, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2PHL
 
 | | THE STRUCTURE OF PHASEOLIN AT 2.2 ANGSTROMS RESOLUTION: IMPLICATIONS FOR A COMMON VICILIN(SLASH)LEGUMIN STRUCTURE AND THE GENETIC ENGINEERING OF SEED STORAGE PROTEINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHASEOLIN, PHOSPHATE ION | | Authors: | Lawrence, M.C, Izard, T, Beuchat, M, Blagrove, R.J, Colman, P.M. | | Deposit date: | 1994-07-07 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of phaseolin at 2.2 A resolution. Implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins.
J.Mol.Biol., 238, 1994
|
|
2WBF
 
 | | Crystal Structure Analysis of SERA5E from plasmodium falciparum with loop 690-700 ordered | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Smith, B.J, Malby, R.L, Colman, P.M, Clarke, O.B. | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the protease-like antigen Plasmodium falciparum SERA5 and its noncanonical active-site serine.
J. Mol. Biol., 392, 2009
|
|
2WH6
 
 | | Crystal structure of anti-apoptotic BHRF1 in complex with the Bim BH3 domain | | Descriptor: | BCL-2-LIKE PROTEIN 11, BROMIDE ION, EARLY ANTIGEN PROTEIN R, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2009-05-01 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1.
PLoS Pathog., 6, 2010
|
|
2XPX
 
 | | Crystal structure of BHRF1:Bak BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BHRF1, BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1.
Plos Pathog., 6, 2010
|
|
2Y6W
 
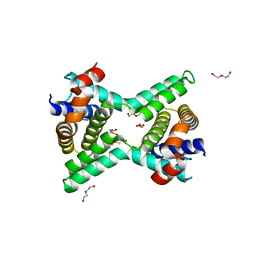 | | Structure of a Bcl-w dimer | | Descriptor: | BCL-2-LIKE PROTEIN 2, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Lee, E.F, Evangelista, M, Pettikiriarachchi, A, Dogovski, C, Perugini, M.A, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2011-01-27 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Bcl-W Domain-Swapped Dimer: Implications for the Function of Bcl-2 Family Proteins.
Structure, 19, 2011
|
|
2YXJ
 
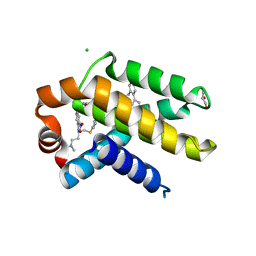 | | Crystal structure of Bcl-xL in complex with ABT-737 | | Descriptor: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, Apoptosis regulator Bcl-X, CHLORIDE ION, ... | | Authors: | Czabotar, P.E, Lee, E.F, Smith, B.J, Deshayes, K, Zobel, K, Fairlie, W.D, Colman, P.M. | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family
Cell Death Differ., 14, 2007
|
|
2YJ1
 
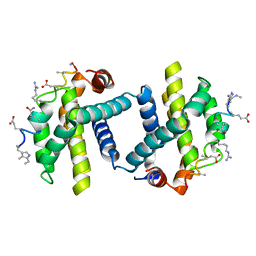 | | Puma BH3 foldamer in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-PUMA BH3 FOLDAMER, BCL-2-LIKE PROTEIN 1 | | Authors: | Lee, E.F, Smith, B.J, Horne, W.S, Mayer, K.N, Evangelista, M, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-05-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Chembiochem, 12, 2011
|
|
1F8C
 
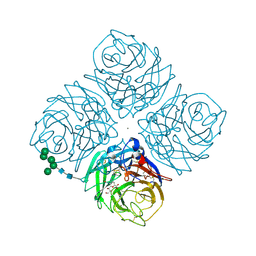 | | Native Influenza Neuraminidase in Complex with 4-amino-2-deoxy-2,3-dehydro-N-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8D
 
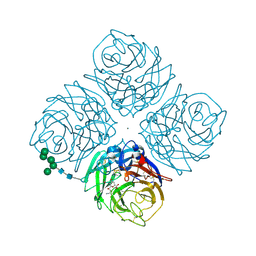 | | Native Influenza Neuraminidase in Complex with 9-amino-2-deoxy-2,3-dehydro-N-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-AMINO-2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8E
 
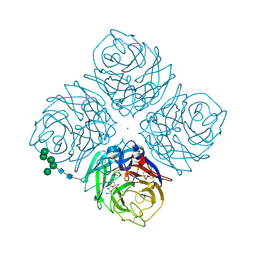 | | Native Influenza Neuraminidase in Complex with 4,9-diamino-2-deoxy-2,3-dehydro-N-acetyl-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,9-AMINO-2,4-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1NMA
 
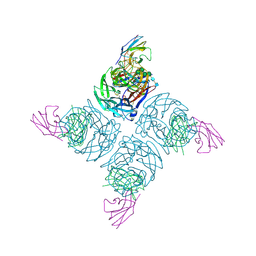 | | N9 NEURAMINIDASE COMPLEXES WITH ANTIBODIES NC41 AND NC10: EMPIRICAL FREE-ENERGY CALCULATIONS CAPTURE SPECIFICITY TRENDS OBSERVED WITH MUTANT BINDING DATA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB NC10, N9 NEURAMINIDASE, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1994-05-06 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N9 neuraminidase complexes with antibodies NC41 and NC10: empirical free energy calculations capture specificity trends observed with mutant binding data.
Biochemistry, 33, 1994
|
|
1PHS
 
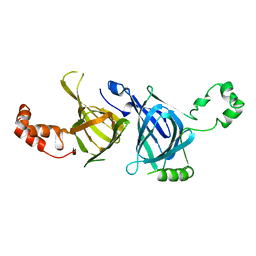 | | THE THREE-DIMENSIONAL STRUCTURE OF THE SEED STORAGE PROTEIN PHASEOLIN AT 3 ANGSTROMS RESOLUTION | | Descriptor: | PHASEOLIN, BETA-TYPE PRECURSOR | | Authors: | Lawrence, M.C, Suzuki, E, Varghese, J.N, Davis, P.C, Vandonkelaar, A, Tulloch, P.A, Colman, P.M. | | Deposit date: | 1990-03-21 | | Release date: | 1990-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The three-dimensional structure of the seed storage protein phaseolin at 3 A resolution.
EMBO J., 9, 1990
|
|
2JBY
 
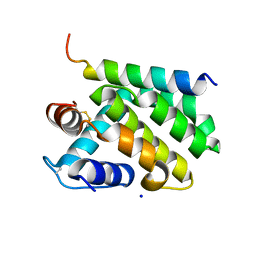 | | A viral protein unexpectedly mimics the structure and function of pro- survival Bcl-2 | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER 2, M11L PROTEIN, SODIUM ION | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak.
Mol. Cell, 25, 2007
|
|
2JBX
 
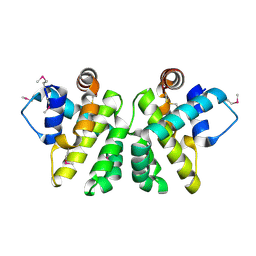 | | Crystal Structure of the myxoma virus anti-apoptotic protein M11L | | Descriptor: | M11L PROTEIN | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Structural Viral Mimic of Prosurvival Bcl-2: A Pivotal Role for Sequestering Proapoptotic Bax and Bak.
Mol.Cell, 25, 2007
|
|
2JM6
 
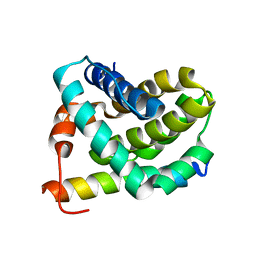 | | Solution structure of MCL-1 complexed with NOXAB | | Descriptor: | Myeloid cell leukemia-1 protein Mcl-1 homolog, Noxa | | Authors: | Czabotar, P.E, Lee, E.F, van Delft, M.F, Day, C.L, Smith, B.J, Huang, D.C.S, Fairlie, W.D, Hinds, M.G, Colman, P.M. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2NLA
 
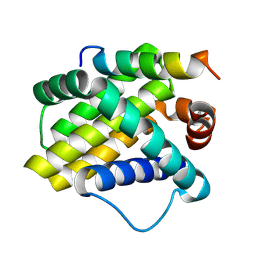 | | Crystal structure of the Mcl-1:mNoxaB BH3 complex | | Descriptor: | FUSION PROTEIN CONSISTING OF Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Phorbol-12-myristate-13-acetate-induced protein 1 | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2006-10-19 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NL9
 
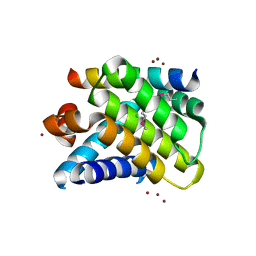 | | Crystal structure of the Mcl-1:Bim BH3 complex | | Descriptor: | Bcl-2-like protein 11, CHLORIDE ION, FUSION PROTEIN CONSISTING OF Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, ... | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2006-10-19 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1NCC
 
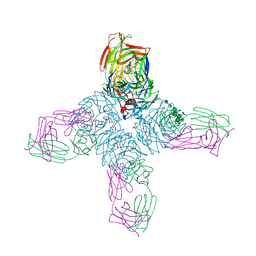 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
1NCD
 
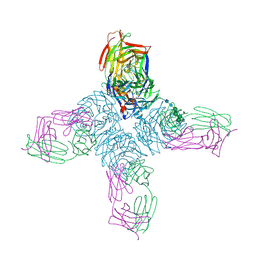 | | REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
J.Mol.Biol., 227, 1992
|
|
1NCA
 
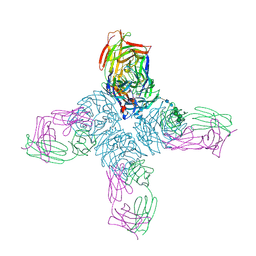 | | REFINED CRYSTAL STRUCTURE OF THE INFLUENZA VIRUS N9 NEURAMINIDASE-NC41 FAB COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex.
J.Mol.Biol., 227, 1992
|
|
1NCB
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
1NMB
 
 | | THE STRUCTURE OF A COMPLEX BETWEEN THE NC10 ANTIBODY AND INFLUENZA VIRUS NEURAMINIDASE AND COMPARISON WITH THE OVERLAPPING BINDING SITE OF THE NC41 ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FAB NC10, ... | | Authors: | Malby, R.L, Tulip, W.R, Colman, P.M. | | Deposit date: | 1995-01-17 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody
Structure, 2, 1994
|
|
3VPP
 
 | | Crystal Structure of the Human CLEC9A C-type Lectin-Like Domain | | Descriptor: | C-type lectin domain family 9 member A, CALCIUM ION | | Authors: | Czabotar, P.E, Zhang, J.G, Policheni, A.N, Shortman, K, Lahoud, M.H, Colman, P.M. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | The dendritic cell receptor Clec9A binds damaged cells via exposed actin filaments.
Immunity, 36, 2012
|
|
3ZLO
 
 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 6) | | Descriptor: | 2-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]-5-(4-phenylbutyl)-1,3-thiazole-4-carboxylic acid, BCL-2-LIKE PROTEIN 1 | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
