7O0Z
 
 | | ABC transporter NosFY, nucleotide-free in GDN | | Descriptor: | Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter permease protein NosY | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7OK4
 
 | |
3FGX
 
 | | Structure of uncharacterised protein rbstp2171 from bacillus stearothermophilus | | Descriptor: | RBSTP2171 | | Authors: | Minasov, G, Filippova, E.V, Shuvalova, L, Moy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Uncharacterised Protein Rbstp2171 from Bacillus Stearothermophilus
To be Published
|
|
6CIU
 
 | | Structure of a Thr-rich interface in an Azami Green tetramer | | Descriptor: | Azami-Green | | Authors: | Oi, C, Lim, C.S, Knecht, K.M, Xiong, Y, Regan, L. | | Deposit date: | 2018-02-25 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A threonine zipper that mediates protein-protein interactions: Structure and prediction.
Protein Sci., 27, 2018
|
|
7O13
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
3FIU
 
 | | Structure of NMN synthetase from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Sorci, L, Martynowski, D, Eyobo, Y, Osterman, A.L, Zhang, H. | | Deposit date: | 2008-12-12 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nicotinamide mononucleotide synthetase is the key enzyme for an alternative route of NAD biosynthesis in Francisella tularensis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3EXZ
 
 | | Crystal structure of the MaoC-like dehydratase from Rhodospirillum rubrum. Northeast Structural Genomics Consortium target RrR103a. | | Descriptor: | MaoC-like dehydratase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Wang, H, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-17 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the MaoC-like dehydratase from Rhodospirillum rubrum.
To be Published
|
|
3F2I
 
 | | Crystal structure of the alr0221 protein from Nostoc, Northeast Structural Genomics Consortium Target NsR422. | | Descriptor: | Alr0221 protein, CHLORIDE ION, PHOSPHATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Belote, R.L, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the alr0221 protein from Nostoc, Northeast Structural Genomics Consortium Target NsR422.
To be Published
|
|
7OQZ
 
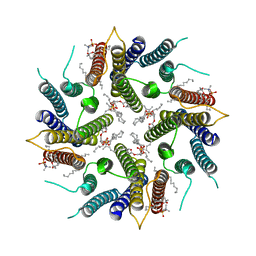 | | Cryo-EM structure of human TMEM45A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Transmembrane protein 45A | | Authors: | Grieben, M, Pike, A.C.W, Evans, A, Shrestha, L, Venkaya, S, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | CryoEM structure of human TMEM45A
To Be Published
|
|
3F2X
 
 | |
3F7O
 
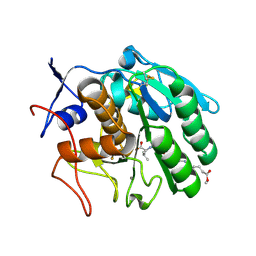 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | Descriptor: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | Authors: | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
3FB2
 
 | | Crystal structure of the human brain alpha spectrin repeats 15 and 16. Northeast Structural Genomics Consortium target HR5563a. | | Descriptor: | Spectrin alpha chain, brain spectrin | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Shastry, R, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human brain alpha spectrin repeats 15 and 16.
To be Published
|
|
1DUS
 
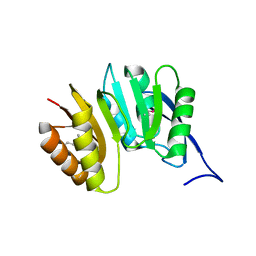 | | MJ0882-A hypothetical protein from M. jannaschii | | Descriptor: | MJ0882 | | Authors: | Hung, L, Huang, L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
3FCB
 
 | |
3FGD
 
 | | Drugscore FP: Thermoylsin in complex with fragment. | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Englert, L, Heine, A, Klebe, G. | | Deposit date: | 2008-12-05 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Drugscore FP: Profiling Protein-Ligand Interactions using Fingerprint Simplicity paired with Knowledge-Based Potential Fields
To be Published
|
|
3FJU
 
 | | Ascaris suum carboxypeptidase inhibitor in complex with human carboxypeptidase A1 | | Descriptor: | ACETATE ION, CACODYLATE ION, Carboxypeptidase A inhibitor, ... | | Authors: | Sanglas, L, Aviles, F.X, Huber, R, Gomis-Ruth, F.X, Arolas, J.L. | | Deposit date: | 2008-12-15 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mammalian metallopeptidase inhibition at the defense barrier of Ascaris parasite
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FN2
 
 | | Crystal structure of a putative sensor histidine kinase domain from Clostridium symbiosum ATCC 14940 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Tesar, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-23 | | Release date: | 2009-01-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of a putative sensor histidine kinase domain from Clostridium symbiosum ATCC 14940
TO BE PUBLISHED
|
|
3FNC
 
 | | Crystal structure of a putative acetyltransferase from Listeria innocua | | Descriptor: | 1,2-ETHANEDIOL, MALONATE ION, Putative acetyltransferase | | Authors: | Cuff, M.E, Tesar, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-24 | | Release date: | 2009-01-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of a putative acetyltransferase from Listeria innocua.
TO BE PUBLISHED
|
|
3FOJ
 
 | | Crystal Structure of SSP1007 From Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Target SyR101A. | | Descriptor: | SODIUM ION, uncharacterized protein | | Authors: | Seetharaman, J, Abashidze, M, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-30 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of SSP1007 From Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Target SyR101A.
To be Published
|
|
3FOM
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide IQQSIERL | | Descriptor: | 8 residue synthetic peptide, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Brims, D.R, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Malashkevich, V.N, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
3FPL
 
 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Felix, F, Goihberg, E, Shimon, L, Burstein, Y. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
3EXA
 
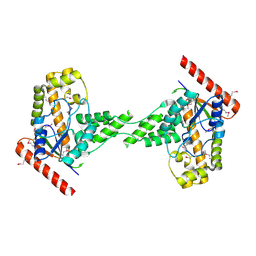 | | Crystal structure of the full-length tRNA isopentenylpyrophosphate transferase (BH2366) from Bacillus halodurans, Northeast Structural Genomics Consortium target BhR41. | | Descriptor: | tRNA delta(2)-isopentenylpyrophosphate transferase | | Authors: | Forouhar, F, Abashidze, M, Neely, H, Seetharaman, J, Shastry, R, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the full-length tRNA isopentenylpyrophosphate transferase (BH2366) from Bacillus halodurans, Northeast Structural Genomics Consortium target BhR41.
To be Published
|
|
3F30
 
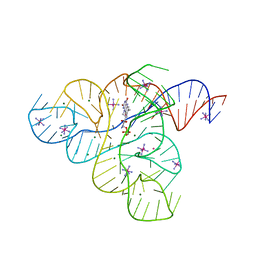 | |
3F2P
 
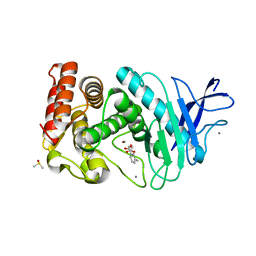 | | Thermolysin inhibition | | Descriptor: | 3-methyl-2-(propanoyloxy)benzoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Englert, L, Heine, A, Klebe, G. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-Based Lead Discovery: Screening and Optimizing Fragments for Thermolysin Inhibition.
Chemmedchem, 5, 2010
|
|
3F2Y
 
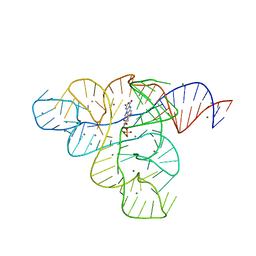 | |
