7E96
 
 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 69% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | Descriptor: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | Authors: | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNW
 
 | | Crystal structure of HpaCas9-sgRNA surveillance complex bound to double-stranded DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand, Target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNT
 
 | |
8H0O
 
 | | Crystal structure of human serum albumin and ruthenium PZA complex adduct | | Descriptor: | Albumin, CHLORIDE ION, NITRIC OXIDE, ... | | Authors: | Gong, W.J, Wang, Y, Bai, H.H, Wang, W.M, Wang, H.F. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Crystal structure of human serum albumin and ruthenium PZA complex adduct
To Be Published
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
6AX3
 
 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
3V71
 
 | | Crystal structure of PUF-6 in complex with 5BE13 RNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Puf (Pumilio/fbf) domain-containing protein 7, confirmed by transcript evidence, ... | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
2JJ3
 
 | | Estrogen receptor beta ligand binding domain in complex with a Benzopyran agonist | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-6-(METHOXYMETHYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, ESTROGEN RECEPTOR BETA | | Authors: | Norman, B.H, Richardson, T.I, Dodge, J.A, Pfeifer, L.A, Durst, G.L, Wang, Y, Durbin, J.D, Krishnan, V, Dinn, S.R, Liu, S, Reilly, J.E, Ryter, K.T. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Benzopyrans as Selective Estrogen Receptor Beta Agonists (Serbas). Part 4: Functionalization of the Benzopyran A-Ring.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8JFU
 
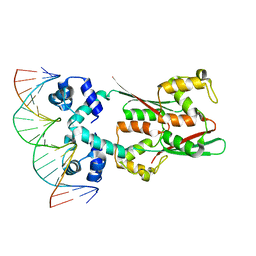 | | AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
6K6I
 
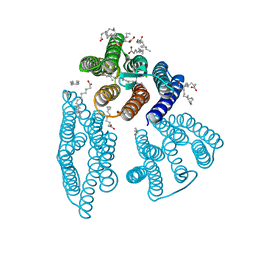 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens
To Be Published
|
|
8JFO
 
 | |
8JFT
 
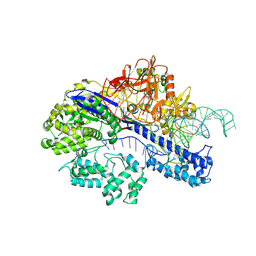 | | Cryo-EM structure of SaCas9-AcrIIA15 CTD-sgRNA complex | | Descriptor: | AcrIIA15, CRISPR-associated endonuclease Cas9, sgRNA of SaCas9 | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFR
 
 | | N-terminal domain of AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JG9
 
 | |
6K6K
 
 | | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens
To Be Published
|
|
6K6J
 
 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion | | Descriptor: | BROMIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion
To Be Published
|
|
3V6Y
 
 | | crystal structure of FBF-2 in complex with a mutant gld-1 FBEa13 RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*AP*CP*UP*GP*UP*GP*CP*CP*AP*UP*AP*C)-3') | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
2KFB
 
 | | The structure of the cataract causing P23T mutant of human gamma-D crystallin | | Descriptor: | Gamma-crystallin D | | Authors: | Jung, J, Byeon, I.L, Wang, Y, King, J, Gronenborn, A.M. | | Deposit date: | 2009-02-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the cataract-causing P23T mutant of human gammaD-crystallin exhibits distinctive local conformational and dynamic changes.
Biochemistry, 48, 2009
|
|
2LF0
 
 | | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering, NESG target SfR339/OCSP target sf3636 | | Descriptor: | Uncharacterized protein yibL | | Authors: | Wu, B, Lemak, A, Yee, A, Lee, H, Gutmanas, A, Semesi, A, Garcia, M, Fang, X, Wang, Y, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering
To be Published
|
|
5ZOX
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 7 at 288 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP9
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 283 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPO
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 8 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PHENYLACETALDEHYDE, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8WRB
 
 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
5ZP6
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 277 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
