8X8S
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WLG
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X8V
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8YV8
 
 | | Cryo-EM structure of CDCA7 bound to nucleosome including hemimethylated CpG site in Widom601 positioning sequence. | | Descriptor: | Cell division cycle-associated protein 7, DNA (132-MER), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, A, Shikimachi, R, Nishiyama, A, Funabiki, H, Arita, K. | | Deposit date: | 2024-03-28 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CDCA7 is a hemimethylated DNA adaptor for the nucleosome remodeler HELLS
To Be Published
|
|
8H78
 
 | | Crystal structure of human MMP-2 catalytic domain in complex with inhibitor | | Descriptor: | (2~{R})-2-[[4-[(4-aminocarbonylphenyl)carbonylamino]phenyl]sulfonylamino]-5-[(2~{S},4~{S})-4-azanyl-2-[[(2~{S})-1-[[(2~{S})-1-[(5-azanyl-5-oxidanylidene-pentyl)amino]-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]-methyl-amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamoyl]pyrrolidin-1-yl]-5-oxidanylidene-pentanoic acid, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Takeuchi, T, Mima, M. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of TP0597850: A Selective, Chemically Stable, and Slow Tight-Binding Matrix Metalloproteinase-2 Inhibitor with a Phenylbenzamide-Pentapeptide Hybrid Scaffold.
J.Med.Chem., 66, 2023
|
|
7DAN
 
 | |
7D4Y
 
 | |
1CNP
 
 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
6NAD
 
 | |
4PLB
 
 | | Crystal Structure of S.A. gyrase-AM8191 complex | | Descriptor: | 6-[({(1r,4S)-1-[(1S)-2-(3-fluoro-6-methoxy-1,5-naphthyridin-4-yl)-1-hydroxyethyl]-2-oxabicyclo[2.2.2]oct-4-yl}amino)methyl]-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, Chimera protein of DNA gyrase subunits B and A, DNA (5'-D(P*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Lu, J, Patel, S, Soisson, S. | | Deposit date: | 2014-05-16 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Oxabicyclooctane-linked novel bacterial topoisomerase inhibitors as broad spectrum antibacterial agents.
Acs Med.Chem.Lett., 5, 2014
|
|
3APS
 
 | | Crystal structure of Trx4 domain of ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10, GLYCEROL, SULFATE ION | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
5H36
 
 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Rhodobacter sphaeroides | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Uncharacterized protein TRIC | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
3CD8
 
 | | X-ray Structure of c-Met with triazolopyridazine Inhibitor. | | Descriptor: | 7-methoxy-4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methoxy]quinoline, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Albrecht, B.K, Harmange, J.-C, Bauer, D, Choquette, D, Dussault, I. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
5H35
 
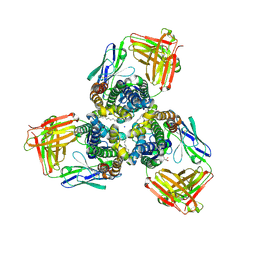 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Sulfolobus solfataricus | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
3CCN
 
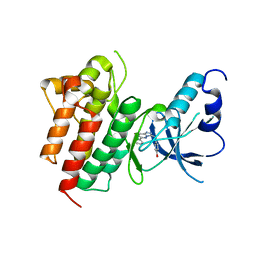 | | X-ray structure of c-Met with triazolopyridazine inhibitor. | | Descriptor: | 4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]phenol, Hepatocyte growth factor receptor | | Authors: | Abrecht, B.K, Harmange, J.-C, Bauer, D, Dussault, I, long, A, Bellon, S.F. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
3I5N
 
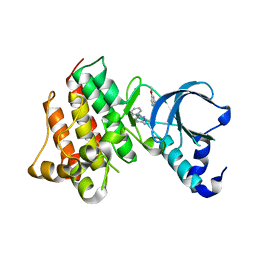 | | Crystal structure of c-Met with triazolopyridazine inhibitor 13 | | Descriptor: | 7-methoxy-N-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]-1,5-naphthyridin-4-amine, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.M, Boezio, A.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of potent and selective triazolopyridazine series of c-Met inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5GHK
 
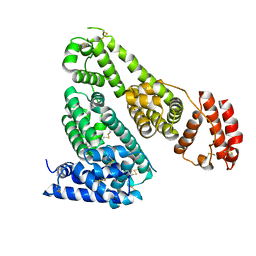 | | Crystal Structure Analysis of Canine serum albumin | | Descriptor: | Serum albumin | | Authors: | Kihira, K, Yamada, K, Kureishi, M, Yokomaku, K, Shinohara, R, Akiyama, M, Komatsu, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Artificial Blood for Dogs
Sci Rep, 6, 2016
|
|
7FH2
 
 | | Crystal structure of the first bromodomain of BRD4 in complex with 16D10 | | Descriptor: | Bromodomain-containing protein 4, CHLORIDE ION, N-[2-[2-[(E)-3-(2,5-dimethoxyphenyl)prop-2-enoyl]-4,5-dimethoxy-phenyl]ethyl]ethanamide | | Authors: | Yokoyama, T, Hirasawa, N. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | A chalcone derivative suppresses TSLP induction in mice and human keratinocytes through binding to BET family proteins.
Biochem Pharmacol, 194, 2021
|
|
5UFO
 
 | | Structure of RORgt bound to | | Descriptor: | (S)-{4-chloro-2-methoxy-3-[4-(methylsulfonyl)phenyl]quinolin-6-yl}(1-methyl-1H-imidazol-5-yl)[6-(trifluoromethyl)pyridin-3-yl]methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UHI
 
 | | Structure of RORgt bound to | | Descriptor: | (R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(4-chlorophenyl)(1-methyl-1H-imidazol-5-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3LC5
 
 | | Selective Benzothiophine Inhibitors of Factor IXa | | Descriptor: | 1-{4-[(R)-phenyl(3-phenyl-1,2,4-oxadiazol-5-yl)methoxy]-1-benzothiophen-2-yl}methanediamine, CALCIUM ION, Coagulation factor IX | | Authors: | Wang, S, Beck, R. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure Based Drug Design: Development of Potent and Selective Factor IXa (FIXa) Inhibitors.
J.Med.Chem., 53, 2010
|
|
5W4R
 
 | | Structure of RORgt bound to a tertiary alcohol | | Descriptor: | 1-{4-[(R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(hydroxy)(1-methyl-1H-imidazol-5-yl)methyl]piperidin-1-yl}ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | 6-Substituted quinolines as ROR gamma t inverse agonists.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5W4V
 
 | | Structure of RORgt bound to a tertiary alcohol | | Descriptor: | (R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(1-methyl-1H-imidazol-5-yl)[6-(trifluoromethyl)pyridin-3-yl]methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Hars, U. | | Deposit date: | 2017-06-13 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 6-Substituted quinolines as ROR gamma t inverse agonists.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6JJU
 
 | | Structure of Ca2+ ATPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Inoue, M, Sakuta, N, Watanabe, S, Inaba, K. | | Deposit date: | 2019-02-27 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Sarco/Endoplasmic Reticulum Ca2+-ATPase 2b Regulation via Transmembrane Helix Interplay.
Cell Rep, 27, 2019
|
|
1A03
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF CA2+-BOUND CALCYCLIN: IMPLICATIONS FOR CA2+-SIGNAL TRANSDUCTION BY S100 PROTEINS, NMR, 20 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, CA2+) | | Authors: | Sastry, M, Ketchem, R.R, Crescenzi, O, Weber, C, Lubienski, M.J, Hidaka, H, Chazin, W.J. | | Deposit date: | 1997-12-08 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of Ca(2+)-bound calcyclin: implications for Ca(2+)-signal transduction by S100 proteins.
Structure, 6, 1998
|
|
