8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
4LST
 
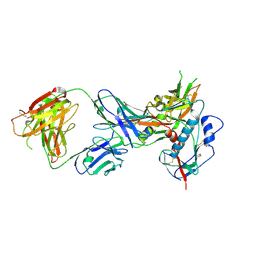 | | Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade C strain ZM176.66 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, HEAVY CHAIN OF ANTIBODY VRC01, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
6E5P
 
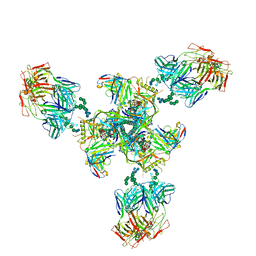 | | Backbone model based on cryo-EM map at 8.5 A of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | | Descriptor: | 2G12 Light chain, 2G12 heavy chain, Envelope glycoprotein gp120, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-07-21 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition.
Structure, 27, 2019
|
|
8D36
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
5MP6
 
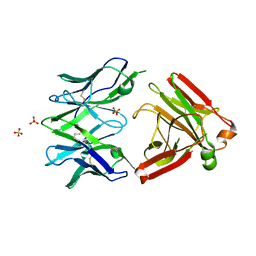 | | Structure of the Unliganded Fab from HIV-1 Neutralizing Antibody CAP248-2B that Binds to the gp120 C-terminus - gp41 Interface, at two Angstrom resolution. | | Descriptor: | CAP248-2B Heavy Chain, CAP248-2B Light Chain, SULFATE ION | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Recognition of a Novel HIV-1 gp120-gp41 Interface Antibody that Caused MPER Exposure through Viral Escape.
PLoS Pathog., 13, 2017
|
|
8D21
 
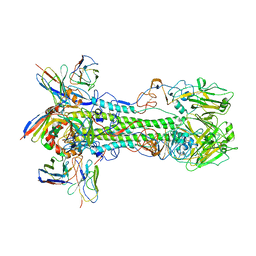 | | Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer | | Descriptor: | 1B06 Heavy Chain, 1B06 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans.
Sci Transl Med, 15, 2023
|
|
7M3I
 
 | |
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
9BEW
 
 | |
9BER
 
 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
9BF6
 
 | |
5WDF
 
 | |
6UBI
 
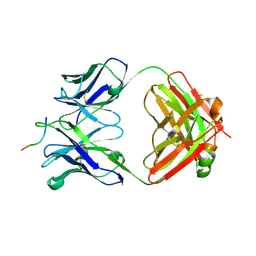 | |
6UDA
 
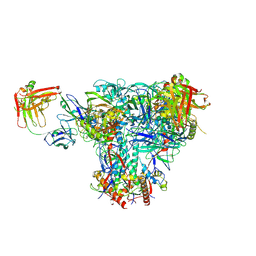 | | Cryo-EM structure of CH235UCA bound to Man5-enriched CH505.N279K.G458Y.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH235 UCA heavy chain Fab, CH235 UCA light chain Fab, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancester of CH235 lineage CD4bs broadly neutralizing antibodies.
Plos Pathog., 15, 2019
|
|
6TYB
 
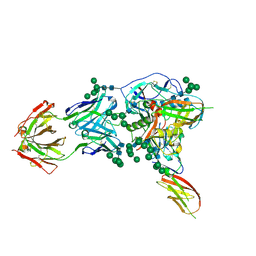 | |
6UCE
 
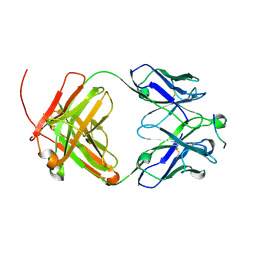 | |
4DKQ
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with DMJ-I-228 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(1S,2S)-2-carbamimidamido-2,3-dihydro-1H-inden-1-yl]-N'-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
4XMP
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC08 in complex with HIV-1 clade A strain Q842.d12 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160,Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-01-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7831 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
8GAS
 
 | | vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-23 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G85
 
 | | vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9W
 
 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
