4ZZI
 
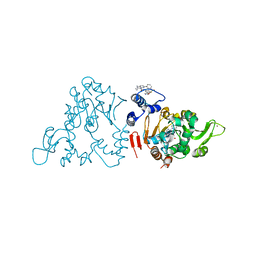 | | SIRT1/Activator/Inhibitor Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7346 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4ZZH
 
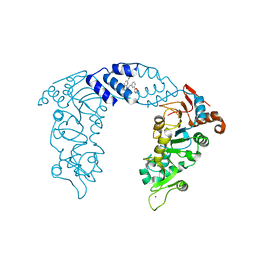 | | SIRT1/Activator Complex | | Descriptor: | (4S)-N-[3-(1,3-oxazol-5-yl)phenyl]-7-[3-(trifluoromethyl)phenyl]-3,4-dihydro-1,4-methanopyrido[2,3-b][1,4]diazepine-5(2H)-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ZINC ION | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
6ZHC
 
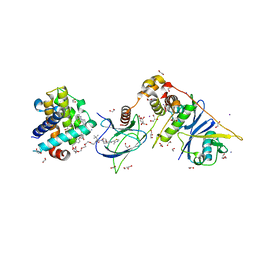 | | PROTAC6 mediated complex of VHL:EloB:EloC and Bcl-xL | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-(1,3-benzothiazol-2-ylcarbamoyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]-5-[3-[4-[3-[2-[2-[2-[2-[2-[3-[[(2~{S})-3,3-dimethyl-1-[(2~{S},4~{R})-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-3-oxidanylidene-propoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]prop-1-ynyl]phenoxy]propyl]-1,3-thiazole-4-carboxylic acid, Bcl-2-like protein 1, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into PROTAC-Mediated Degradation of Bcl-xL.
Acs Chem.Biol., 15, 2020
|
|
7R8V
 
 | | Cryo-EM structure of the ADP state actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Espinosa de los Reyes, S, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-06-27 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7RB9
 
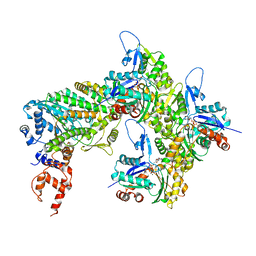 | | Cryo-EM structure of the rigor state Jordan myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7R91
 
 | | cryo-EM structure of the rigor state wild type myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7RB8
 
 | | cryo-EM structure of the ADP state wild type myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
6R7D
 
 | | Crystal structure of LTC4S in complex with AZ13690257 | | Descriptor: | (1~{S},2~{S})-2-[5-[cyclopropylmethyl(naphthalen-1-yl)amino]-4-methoxy-pyrimidin-2-yl]carbonylcyclopropane-1-carboxylic acid, DODECYL-BETA-D-MALTOSIDE, Leukotriene C4 synthase, ... | | Authors: | Kack, H, Ek, M. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of the Oral Leukotriene C4 Synthase Inhibitor (1S,2S)-2-({5-[(5-Chloro-2,4-difluorophenyl)(2-fluoro-2-methylpropyl)amino]-3-methoxypyrazin-2-yl}carbonyl)cyclopropanecarboxylic Acid (AZD9898) as a New Treatment for Asthma.
J.Med.Chem., 62, 2019
|
|
5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
6TXZ
 
 | | FAB PART OF M6903 IN COMPLEX WITH HUMAN TIM3 | | Descriptor: | Fab H, Fab L, Hepatitis A virus cellular receptor 2 | | Authors: | Musil, D, Sood, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Identification and characterization of M6903, an antagonistic anti-TIM-3 monoclonal antibody.
Oncoimmunology, 9, 2020
|
|
1J4J
 
 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
1MOL
 
 | |
4FVT
 
 | | Human SIRT3 bound to Ac-ACS peptide and Carba-NAD | | Descriptor: | Acetylated ACS2 peptide, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dai, H. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-15 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Synthesis of Carba-NAD and the Structures of Its Ternary Complexes with SIRT3 and SIRT5.
J.Org.Chem., 77, 2012
|
|
4G1N
 
 | | PKM2 in complex with an activator | | Descriptor: | MAGNESIUM ION, N-(4-{[4-(pyrazin-2-yl)piperazin-1-yl]carbonyl}phenyl)quinoline-8-sulfonamide, OXALATE ION, ... | | Authors: | Kung, C, Hixon, J, Dang, L, DeLaBarre, B, Qian, K.C. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small Molecule Activation of PKM2 in Cancer Cells Induces Serine Auxotrophy.
Chem.Biol., 19, 2012
|
|
4G1C
 
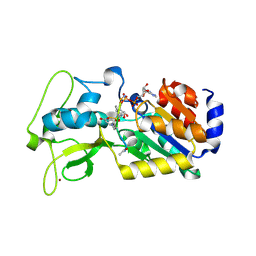 | | Human SIRT5 bound to Succ-IDH2 and Carba-NAD | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-dependent protein deacylase sirtuin-5, mitochondrial, ... | | Authors: | Dai, H. | | Deposit date: | 2012-07-10 | | Release date: | 2012-08-15 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Synthesis of Carba-NAD and the Structures of Its Ternary Complexes with SIRT3 and SIRT5.
J.Org.Chem., 77, 2012
|
|
5IZ8
 
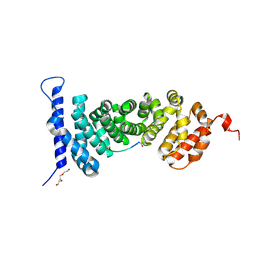 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ9
 
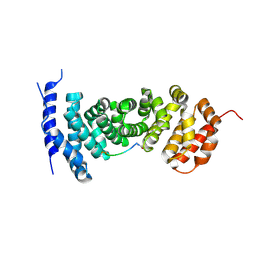 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZA
 
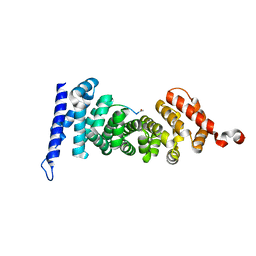 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ6
 
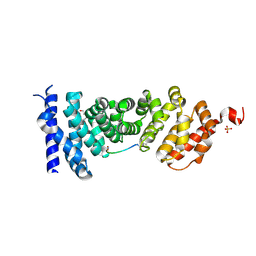 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
8V7Z
 
 | |
8V81
 
 | | Phosphorylated, ATP-bound, inhibitor 172-bound E1371Q human cystic fibrosis transmembrane conductance regulator | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(Z)-{(3M)-4-oxo-2-sulfanylidene-3-[3-(trifluoromethyl)phenyl]-1,3-thiazolidin-5-ylidene}methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gao, X, Hwang, T. | | Deposit date: | 2023-12-04 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric inhibition of CFTR gating by CFTRinh-172 binding in the pore
Nat Commun, 2024
|
|
4JA8
 
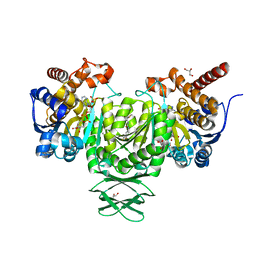 | | Complex of Mitochondrial Isocitrate Dehydrogenase R140Q Mutant with AGI-6780 Inhibitor | | Descriptor: | 1-[5-(cyclopropylsulfamoyl)-2-thiophen-3-yl-phenyl]-3-[3-(trifluoromethyl)phenyl]urea, CALCIUM ION, GLYCEROL, ... | | Authors: | Wei, W, Chen, L, Wu, M, Jiang, F, Travins, J, Qian, K, DeLaBarre, B. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation.
Science, 340, 2013
|
|
5TBP
 
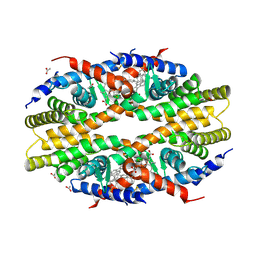 | | Crystal Structure of RXR-alpha ligand binding domain complexed with synthetic modulator K8003 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Aleshin, A.E, Liddington, R.C, Su, Y, Zhang, X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modulation of nongenomic activation of PI3K signalling by tetramerization of N-terminally-cleaved RXR alpha.
Nat Commun, 8, 2017
|
|
4N5G
 
 | | Crystal Structure of RXRa LBD complexed with a synthetic modulator K8012 | | Descriptor: | 5-(2-{(1Z)-5-fluoro-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
4N8R
 
 | | Crystal structure of RXRa LBD complexed with a synthetic modulator K-8008 | | Descriptor: | 5-(2-{(1Z)-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
