4O1D
 
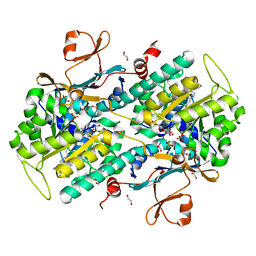 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O15
 
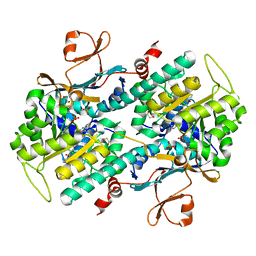 | | The crystal structure of a mutant NAMPT (S165F) in complex with GNE-618 | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O16
 
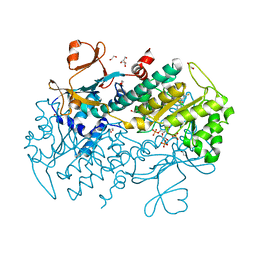 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 6-({4-[(3,5-difluorophenyl)sulfonyl]benzyl}carbamoyl)-1-(5-O-phosphono-beta-D-ribofuranosyl)imidazo[1,2-a]pyridin-1-ium, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7C00
 
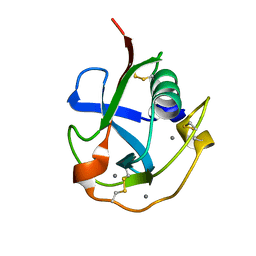 | | Crystal structure of the SRCR domain of human SCARA5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor class A member 5 | | Authors: | Yu, B, He, Y. | | Deposit date: | 2020-04-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of ferritin with scavenger receptor class A members.
J.Biol.Chem., 295, 2020
|
|
7BZZ
 
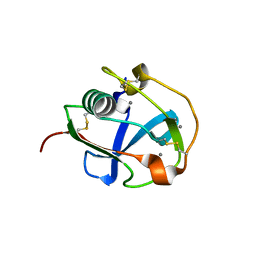 | | Crystal structure of the SRCR domain of mouse SCARA5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor class A member 5 | | Authors: | Yu, B, He, Y. | | Deposit date: | 2020-04-29 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Interactions of ferritin with scavenger receptor class A members.
J.Biol.Chem., 295, 2020
|
|
6UBI
 
 | |
6UCE
 
 | |
6UCF
 
 | |
6XWE
 
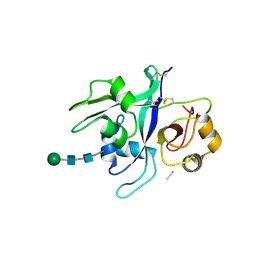 | | Crystal structure of LYK3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-01-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
7DPX
 
 | |
3SBE
 
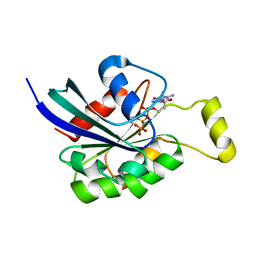 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
1FM4
 
 | | CRYSTAL STRUCTURE OF THE BIRCH POLLEN ALLERGEN BET V 1L | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, MAJOR POLLEN ALLERGEN BET V 1-L | | Authors: | Markovic-Housley, Z, Degano, M, Lamba, D, von Roepenack-Lahaye, E, Clemens, S, Susani, M, Ferreira, F, Scheiner, O, Breiteneder, H. | | Deposit date: | 2000-08-16 | | Release date: | 2002-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a Hypoallergenic Isoform of the Major Birch Pollen Allergen Bet
v 1 and its Likely Biological Function as a Plant Steroid Carrier
J.Mol.Biol., 325, 2003
|
|
2BIT
 
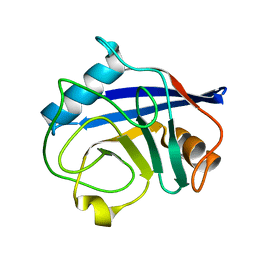 | | Crystal structure of human cyclophilin D at 1.7 A resolution | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BIU
 
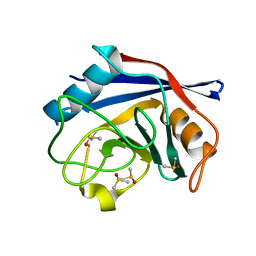 | | Crystal structure of human cyclophilin D at 1.7 A resolution, DMSO complex | | Descriptor: | DIMETHYL SULFOXIDE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7KJS
 
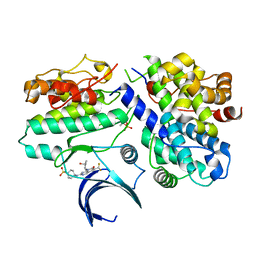 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | Descriptor: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | McTigue, M.A, He, Y, Ferre, R.A. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
2AID
 
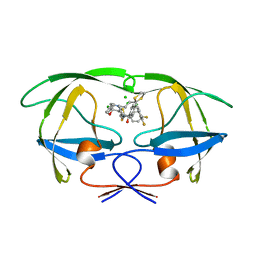 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
1AID
 
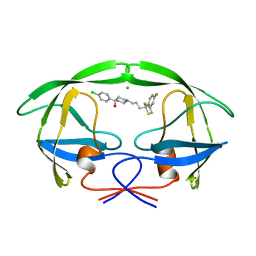 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
8SXS
 
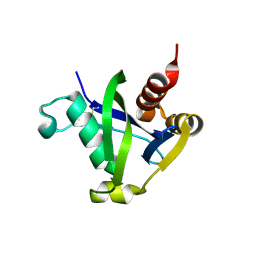 | |
7RBS
 
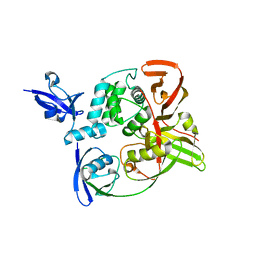 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with human ISG15 | | Descriptor: | Papain-like protease, Ubiquitin-like protein ISG15, ZINC ION | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7RBR
 
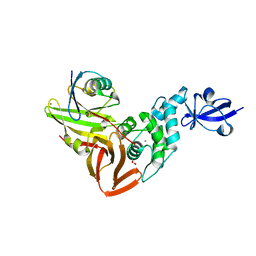 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Papain-like protease, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7S6O
 
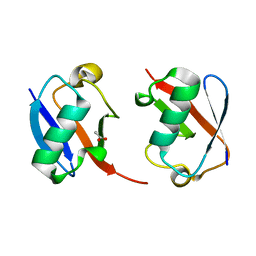 | | The crystal structure of Lys48-linked di-ubiquitin | | Descriptor: | ACETATE ION, Ubiquitin | | Authors: | Osipiuk, J, Tesar, C, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7S6P
 
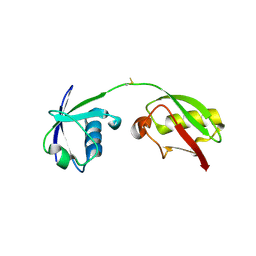 | | The crystal structure of human ISG15 | | Descriptor: | Ubiquitin-like protein ISG15 | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
5AQ0
 
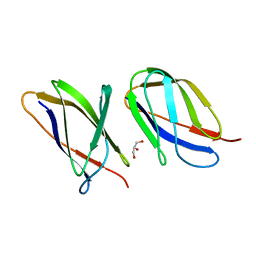 | | The structure of the Transthyretin-like domain of the first catalytic domain of the HUMAN Carboxypeptidase D | | Descriptor: | CARBOXYPEPTIDASE D, GLYCEROL | | Authors: | Gallego, P, Garcia-Pardo, J, Lorenzo, J, Aviles, F.X, Ventura, S, Reverter, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The Structure of the Ttldomain of the Human Carboxypeptidase D
To be Published
|
|
4JNA
 
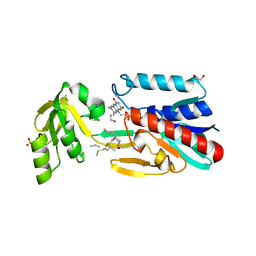 | | Crystal structure of the DepH complex with dimethyl-FK228 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | Authors: | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
