5MGI
 
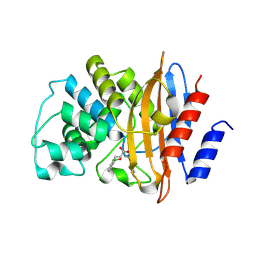 | | Crystal structure of KPC-2 carbapenemase in complex with a phenyl boronic inhibitor. | | Descriptor: | (~{E})-3-[2-(dihydroxyboranyl)phenyl]prop-2-enoic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Vicario, M, Celenza, G, Bellio, P, Perilli, M.G, Tondi, D, Cendron, L. | | Deposit date: | 2016-11-21 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phenylboronic Acid Derivatives as Validated Leads Active in Clinical Strains Overexpressing KPC-2: A Step against Bacterial Resistance.
Chemmedchem, 13, 2018
|
|
1ZOF
 
 | | Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter Pylori | | Descriptor: | alkyl hydroperoxide-reductase | | Authors: | Papinutto, E, Windle, H.J, Cendron, L, Battistutta, R, Kelleher, D, Zanotti, G. | | Deposit date: | 2005-05-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter pylori.
Biochim.Biophys.Acta, 1753, 2005
|
|
2NOY
 
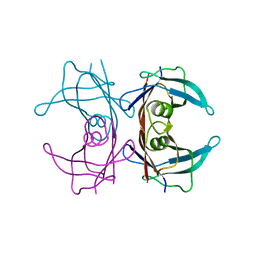 | | Crystal structure of transthyretin mutant I84S at PH 7.5 | | Descriptor: | Transthyretin | | Authors: | Pasquato, N, Berni, R, Folli, C, Alfieri, B, Cendron, L, Zanotti, G. | | Deposit date: | 2006-10-26 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acidic pH-induced conformational changes in amyloidogenic mutant transthyretin
J.Mol.Biol., 366, 2007
|
|
7ZRA
 
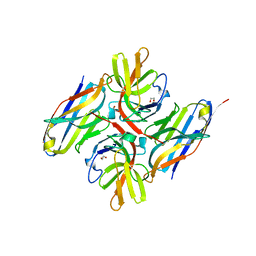 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS1(Nb14497) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody NbSOS1 (Nb14497) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2022-05-04 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
6TGI
 
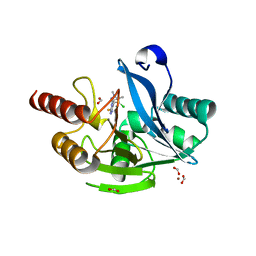 | | Crystal structure of VIM-2 in complex with triazole-based inhibitor OP24 | | Descriptor: | 5-(4-chloranyl-1,5-dimethyl-pyrazol-3-yl)-4-ethyl-1,2,4-triazole-3-thiol, FORMIC ACID, Vim-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
8PDE
 
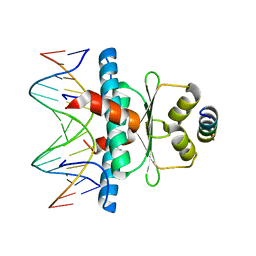 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC4 deacetylase binding motif | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), HDAC4 (histone deacetylase 4) binding motif peptide:GSGEVKMKLQEFVLNKK, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-06-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
6TS9
 
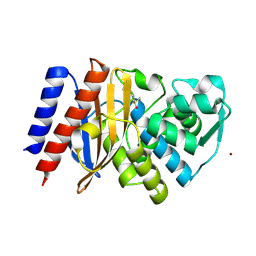 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-lactamase, ... | | Authors: | Maso, L, Tondi, D, Klein, R, Montanari, M, Bellio, C, Celenza, G, Brenk, R, Cendron, L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the Class A Carbapenemase GES-5 via Virtual Screening.
Biomolecules, 10, 2020
|
|
8Q9R
 
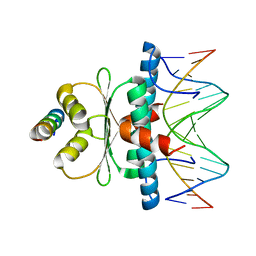 | | Crystal structure of MADS-box/MEF2D N-terminal domain bound to dsDNA and HDAC9 deacetylase binding motif | | Descriptor: | Histone deacetylase 9 (HDAC9) binding motif peptide: EVKQKLQEFLLSKS, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8Q9P
 
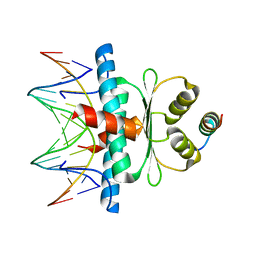 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC5 deacetylase binding motif | | Descriptor: | HDAC5 (histone deacetylase 5) binding motif peptide: TRP-GLY-SER-GLY-GLU-VAL-LYS-LEU-ARG-LEU-GLN-GLU-PHE-LEU-LEU-SER-LYS-SER, MADS box dsDNA fw: AACTATTTATAAGA, MADS box dsNA rev:TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8Q9Q
 
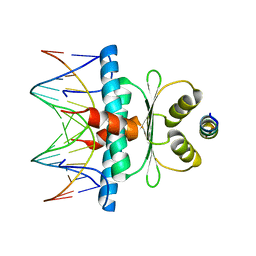 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC7 deacetylase binding motif | | Descriptor: | HDAC7 (histone deacetylase 7) binding motif peptide: GLY-VAL-VAL-LYS-GLN-LYS-LEU-ALA-GLU-VAL-ILE-LEU-LYS-LYS-GLN, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8Q9N
 
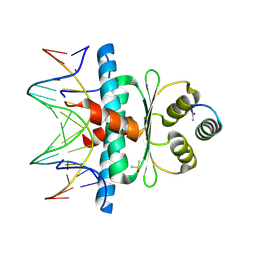 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and MITR deacetylase binding motif mutant L151V. | | Descriptor: | DIMETHYL SULFOXIDE, DNA MADS box, MEF2D protein, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8B20
 
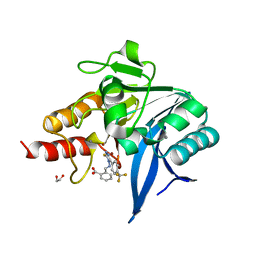 | | NDM-1 metallo-beta-lactamase in complex with triazole-based inhibitor CP57 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(~{E})-[3-sulfanyl-5-[4-(trifluoromethyl)phenyl]-1,2,4-triazol-4-yl]iminomethyl]benzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Vascon, F, Lazzarato, L, Bersani, M, Gianquinto, E, Docquier, J.D, Spyrakis, F, Tondi, D, Cendron, L. | | Deposit date: | 2022-09-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Optimization of 1,2,4-Triazole-3-Thione Derivatives: Improving Inhibition of NDM-/VIM-Type Metallo-beta-Lactamases and Synergistic Activity on Resistant Bacteria.
Pharmaceuticals, 16, 2023
|
|
8B1Z
 
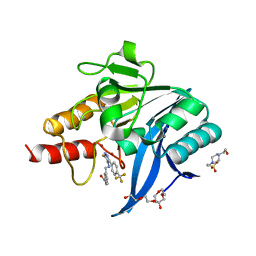 | | NDM-1 metallo-beta-lactamase in complex with triazole-based inhibitor CP56 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-[3-sulfanyl-5-[4-(trifluoromethyl)phenyl]-1,2,4-triazol-4-yl]iminomethyl]benzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Vascon, F, Lazzarato, L, Bersani, M, Gianquinto, E, Docquier, J.D, Spyrakis, F, Tondi, D, Cendron, L. | | Deposit date: | 2022-09-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of 1,2,4-Triazole-3-Thione Derivatives: Improving Inhibition of NDM-/VIM-Type Metallo-beta-Lactamases and Synergistic Activity on Resistant Bacteria.
Pharmaceuticals, 16, 2023
|
|
8B1W
 
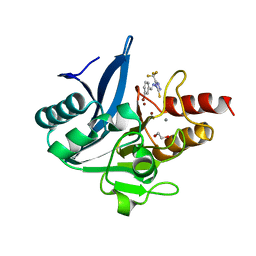 | | NDM-1 metallo-beta-lactamase in complex with triazole-based inhibitor CP35 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[(~{E})-(3-bromophenyl)methylideneamino]-5-(trifluoromethyl)-1,2,4-triazole-3-thiol, ... | | Authors: | Vascon, F, Lazzarato, L, Bersani, M, Gianquinto, E, Docquier, J.D, Spyrakis, F, Tondi, D, Cendron, L. | | Deposit date: | 2022-09-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Optimization of 1,2,4-Triazole-3-Thione Derivatives: Improving Inhibition of NDM-/VIM-Type Metallo-beta-Lactamases and Synergistic Activity on Resistant Bacteria.
Pharmaceuticals, 16, 2023
|
|
7Z57
 
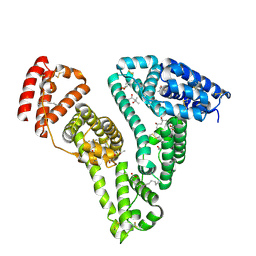 | | Crystal structure of Human Serum Albumin in complex with surfactant GenX (2,3,3,3-tetrafluoro-2-(heptafluoropropoxy) propanoate) | | Descriptor: | (2R)-2,3,3,3-tetrakis(fluoranyl)-2-[1,1,2,2,3,3,3-heptakis(fluoranyl)propoxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Albumin, ... | | Authors: | Liberi, S, Moro, G, Vascon, F, Linciano, S, De Toni, L, Angelini, A, Cendron, L. | | Deposit date: | 2022-03-08 | | Release date: | 2022-10-12 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigation of the Interaction between Human Serum Albumin and Branched Short-Chain Perfluoroalkyl Compounds.
Chem.Res.Toxicol., 35, 2022
|
|
8A76
 
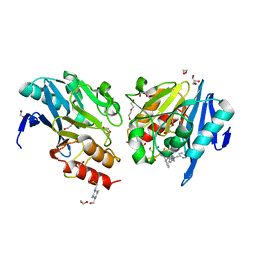 | | Metallo-beta-lactamase NDM-1 in complex with 1,2,4-Triazole-3-thione compound 26 | | Descriptor: | (2~{S})-2-[bis(pyridin-2-ylmethyl)amino]-5-(3-phenyl-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl)pentanoic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Vascon, F, Legru, A, Hernandez, J.F, Cendron, L. | | Deposit date: | 2022-06-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of 1,2,4-Triazole-3-thiones toward Broad-Spectrum Metallo-beta-lactamase Inhibitors Showing Potent Synergistic Activity on VIM- and NDM-1-Producing Clinical Isolates.
J.Med.Chem., 65, 2022
|
|
8C84
 
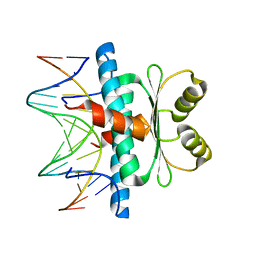 | | Crystal structure of MADS-box/MEF2D N-terminal domain complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), MEF2D protein | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
4ZZF
 
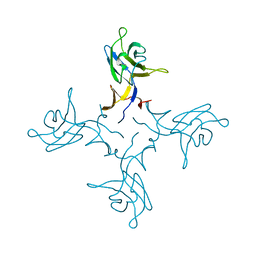 | | Crystal structure of truncated FlgD (tetragonal form) from the human pathogen Helicobacter pylori | | Descriptor: | Flagellar basal body rod modification protein | | Authors: | Pulic, I, Cendron, L, Salamina, M, Matkovic-Calogovic, D, Zanotti, G. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1673 Å) | | Cite: | Crystal structure of truncated FlgD from the human pathogen Helicobacter pylori.
J.Struct.Biol., 194, 2016
|
|
4ZZK
 
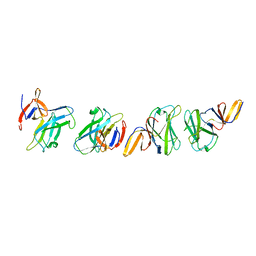 | | Crystal structure of truncated FlgD (monoclinic form) from the human pathogen Helicobacter pylori | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Pulic, I, Cendron, L, Salamina, M, Matkovic-Calogovic, D, Zanotti, G. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of truncated FlgD from the human pathogen Helicobacter pylori.
J.Struct.Biol., 194, 2016
|
|
3BT0
 
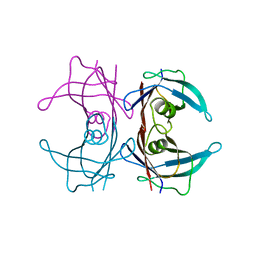 | | Crystal structure of transthyretin variant V20S | | Descriptor: | Transthyretin | | Authors: | Zanotti, G, Folli, C, Cendron, L, Gliubich, F, Negro, A, Berni, R. | | Deposit date: | 2007-12-27 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
7AAE
 
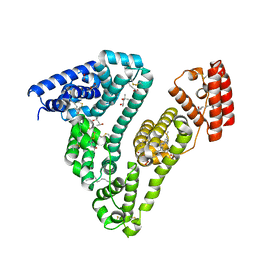 | | Crystal structure of Human serum albumin in complex with myristic acid at 2.27 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Albumin, FORMIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
7AAI
 
 | | Crystal structure of Human serum albumin in complex with perfluorooctanoic acid (PFOA) at 2.10 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
7ZRR
 
 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, AMINO GROUP, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965
To Be Published
|
|
7ZRT
 
 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970
To Be Published
|
|
6T8C
 
 | | Crystal structure of formate dehydrogenase FDH2 enzyme from Granulicella mallensis MP5ACTX8 in the apo form. | | Descriptor: | Formate dehydrogenase | | Authors: | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | From the Amelioration of a NADP+-dependent Formate Dehydrogenase to the Discovery of a New Enzyme: Round Trip from Theory to Practice
Chemcatchem, 2020
|
|
