4JZB
 
 | | Crystal Structure of Leshmaniasis major Farnesyl diphosphate synthase in complex with 1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)-3-PHENYLPYRIDINIUM, IPP and Ca2+ | | Descriptor: | 1-(2-hydroxy-2,2-diphosphonoethyl)-3-phenylpyridinium, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S. | | Deposit date: | 2013-04-02 | | Release date: | 2014-02-12 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic basis of the inhibition of Leishmania major farnesyl diphosphate synthase by nitrogen-containing bisphosphonates.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2BRA
 
 | | Structure of N-Terminal FAD Binding motif of mouse MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9 INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Nadella, M, Bianchet, M.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the axon guidance protein MICAL.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
4JZX
 
 | | Crystal Structure of Leshmaniasis major Farnesyl diphosphate synthase in complex with 3-BUTYL-1-(2,2-DIPHOSPHONOETHYL)PYRIDINIUM, IPP and Ca2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, CALCIUM ION, ... | | Authors: | Aripirala, S, Gabelli, S, Amzel, L.M. | | Deposit date: | 2013-04-03 | | Release date: | 2014-03-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic basis of the inhibition of Leishmania major farnesyl diphosphate synthase by nitrogen-containing bisphosphonates.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4K10
 
 | | Crystal Structure of Leshmaniasis major Farnesyl diphosphate synthase in complex with 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM and Mg2+ | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Aripirala, S, Gabelli, S, Amzel, L.M. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic basis of the inhibition of Leishmania major farnesyl diphosphate synthase by nitrogen-containing bisphosphonates.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2HXM
 
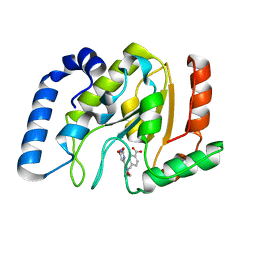 | | Complex of UNG2 and a small Molecule synthetic Inhibitor | | Descriptor: | 4-[(1E,7E)-8-(2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDIN-4-YL)-3,6-DIOXA-2,7-DIAZAOCTA-1,7-DIEN-1-YL]BENZOIC ACID, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Ghung, S, Seiple, L, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mimicking damaged DNA with a small molecule inhibitor of human UNG2.
Nucleic Acids Res., 34, 2006
|
|
2GT2
 
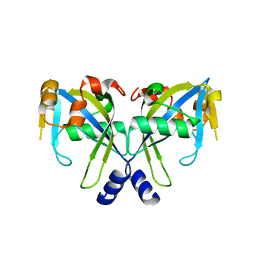 | | Structure of the E. coli GDP-mannose mannosyl hydrolase | | Descriptor: | GDP-mannose mannosyl hydrolase | | Authors: | Gabelli, S.B, Bianchet, M.A, Azurmendi, H.F, MIldvan, A.S, Amzel, L.M. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, NMR, and mutational studies of the catalytic cycle of the GDP-mannose mannosyl hydrolase reaction.
Biochemistry, 45, 2006
|
|
2GSI
 
 | |
7UUZ
 
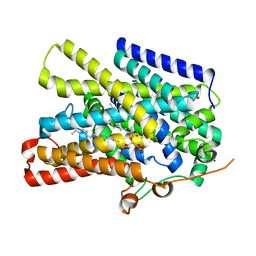 | | Structure of the sodium/iodide symporter (NIS) in complex with perrhenate and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, PERRHENATE, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
7UUY
 
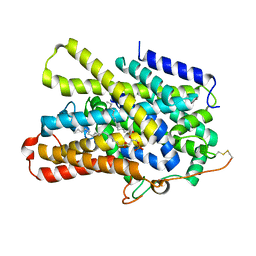 | | Structure of the sodium/iodide symporter (NIS) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Sodium/iodide cotransporter | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
7UV0
 
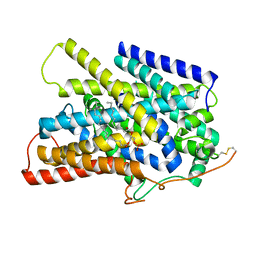 | | Structure of the sodium/iodide symporter (NIS) in complex with iodide and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, IODIDE ION, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
4OVN
 
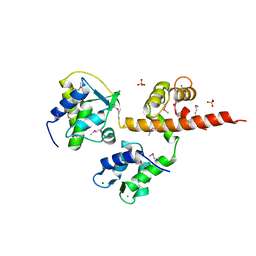 | | Voltage-gated Sodium Channel 1.5 (Nav1.5) C-terminal domain in complex with Calmodulin poised for activation | | Descriptor: | Calmodulin, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Boto, A, Jakoncic, J, Tomaselli, G.F, Amzel, L.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of the NaV1.5 cytoplasmic domain by calmodulin.
Nat Commun, 5, 2014
|
|
4OVV
 
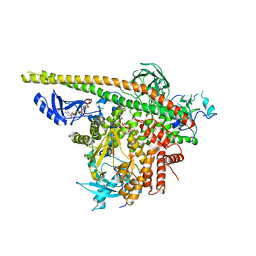 | | Crystal Structure of PI3Kalpha in complex with diC4-PIP2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
4OVU
 
 | | Crystal Structure of p110alpha in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
1GA7
 
 | | CRYSTAL STRUCTURE OF THE ADP-RIBOSE PYROPHOSPHATASE IN COMPLEX WITH GD+3 | | Descriptor: | GADOLINIUM ION, HYPOTHETICAL 23.7 KDA PROTEIN IN ICC-TOLC INTERGENIC REGION | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2000-11-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family.
Nat.Struct.Biol., 8, 2001
|
|
1HDY
 
 | |
1HDZ
 
 | |
1HDX
 
 | |
1RYA
 
 | | Crystal Structure of the E. coli GDP-mannose mannosyl hydrolase in complex with GDP and MG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GDP-mannose mannosyl hydrolase, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Legler, P.M, Mildvan, A.S, Amzel, L.M. | | Deposit date: | 2003-12-20 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of GDP-mannose glycosyl hydrolase, a Nudix enzyme that cleaves at carbon instead of phosphorus.
Structure, 12, 2004
|
|
6NCT
 
 | | Structure of p110alpha/niSH2 - vector data collection | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION, ... | | Authors: | Miller, M.S, Maheshwari, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
1SDW
 
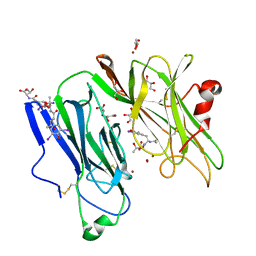 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase with bound peptide and dioxygen | | Descriptor: | COPPER (II) ION, GLYCEROL, N-ALPHA-ACETYL-3,5-DIIODOTYROSYL-D-THREONINE, ... | | Authors: | Prigge, S.T, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dioxygen binds end-on to mononuclear copper in a precatalytic enzyme complex.
Science, 304, 2004
|
|
6MC9
 
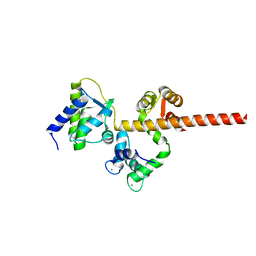 | |
3Q1P
 
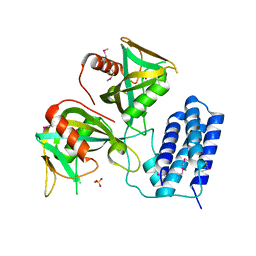 | | Crystal structure of CDP-Chase | | Descriptor: | Phosphohydrolase (MutT/nudix family protein), SULFATE ION | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-17 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
5C7Q
 
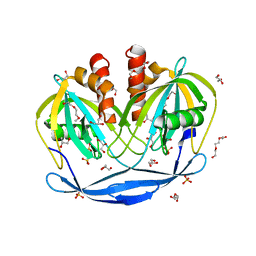 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
5C7T
 
 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase in complex with ADP-ribose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5-DIPHOSPHORIBOSE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
1CK0
 
 | |
