8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | Descriptor: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | Descriptor: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | Descriptor: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7CRH
 
 | | Cryo-EM structure of SKF83959 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1S)-6-chloranyl-3-methyl-1-(3-methylphenyl)-1,2,4,5-tetrahydro-3-benzazepine-7,8-diol, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yan, W, Shao, Z.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKW
 
 | | Cryo-EM structure of Fenoldopam bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKZ
 
 | | Cryo-EM structure of Dopamine and LY3154207 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKY
 
 | | Cryo-EM structure of PW0464 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 6-[4-[3-[bis(fluoranyl)methoxy]pyridin-2-yl]oxy-2-methyl-phenyl]-1,5-dimethyl-pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKX
 
 | | Cryo-EM structure of A77636 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R,3S)-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1H-isochromene-5,6-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
4X7Q
 
 | | PIM2 kinase in complex with Compound 1s | | Descriptor: | 2-(2,6-difluorophenyl)-N-{4-[(3S)-pyrrolidin-3-yloxy]pyridin-3-yl}-1,3-thiazole-4-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-2 | | Authors: | Marcotte, D.J, Silvian, L.F. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-based design of low-nanomolar PIM kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3LPI
 
 | | Structure of BACE Bound to SCH745132 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
5H5Z
 
 | | Crystal structure of bony fish MHC class I, peptide and B2m II | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide chain | | Authors: | Chen, Z, Zhang, N, Qi, J, Li, X, Chen, R, Wang, Z, Gao, F.G, Xia, C. | | Deposit date: | 2016-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Mechanism of beta 2m Molecule-Induced Changes in the Peptide Presentation Profile in a Bony Fish.
Iscience, 23, 2020
|
|
3LPK
 
 | | Structure of BACE Bound to SCH747123 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5DZN
 
 | | human T-cell immunoglobulin and mucin domain protein 4 | | Descriptor: | T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Yuan, S, Rao, Z, Wang, X. | | Deposit date: | 2015-09-25 | | Release date: | 2015-11-25 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope.
Protein Cell, 6, 2015
|
|
3LNK
 
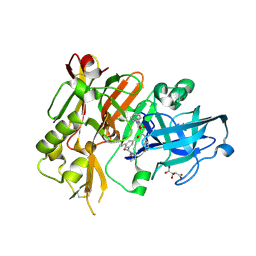 | | Structure of BACE bound to SCH743813 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Orth, P, Cumming, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPJ
 
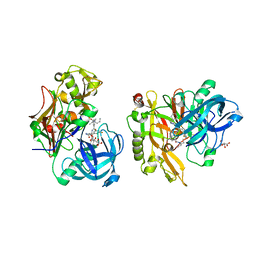 | | Structure of BACE Bound to SCH743641 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-[(1S,2S)-2-[(2R)-4-benzylpiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7ZYJ
 
 | |
6AJI
 
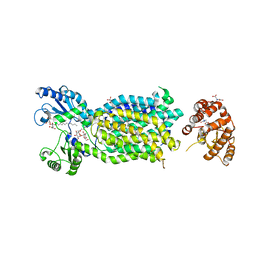 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with Rimonabant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
5ILB
 
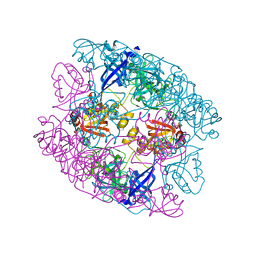 | | Crystal structure of protease domain of Deg2 linked with the PDZ domain of Deg9 | | Descriptor: | Protease Do-like 2, chloroplastic,Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
6AJF
 
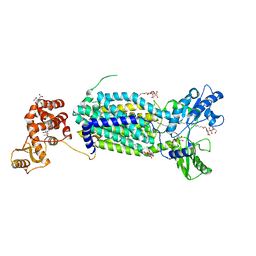 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Drug exporters of the RND superfamily-like protein,Endolysin, alpha-D-glucopyranosyl 6-O-dodecyl-alpha-D-glucopyranoside | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJG
 
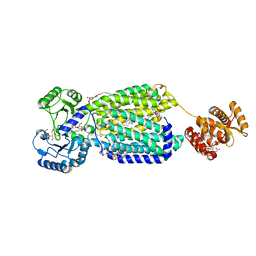 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SQ109 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, DODECYL-BETA-D-MALTOSIDE, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJJ
 
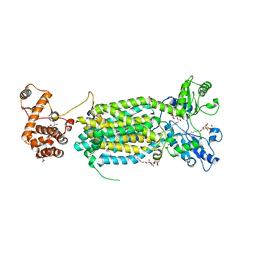 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with ICA38 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4,6-difluoro-N-(spiro[5.5]undecan-3-yl)-1H-indole-2-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
5IL9
 
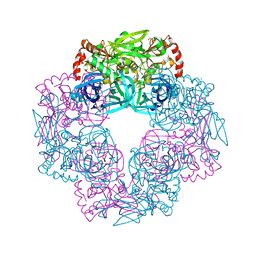 | | Crystal structure of Deg9 | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
8IZE
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 4-HMBPP | | Descriptor: | Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, [(E)-3-(hydroxymethyl)pent-2-enyl] phosphono hydrogen phosphate | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
