7L5D
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with demethylated analog of masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
6DWE
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate- and BRD0059-bound form | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-26 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
6DVV
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | Descriptor: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
1L1S
 
 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
6DXN
 
 | | 1.95 Angstrom Resolution Crystal Structure of DsbA Disulfide Interchange Protein from Klebsiella pneumoniae. | | Descriptor: | TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
5D8M
 
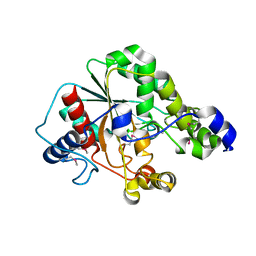 | | Crystal structure of the metagenomic carboxyl esterase MGS0156 | | Descriptor: | Metagenomic carboxyl esterase MGS0156 | | Authors: | Cui, H, Nocek, B, Tchigvintsev, A, Popovic, A, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2015-08-17 | | Release date: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of esterase (MGS0156)
To Be Published
|
|
1L0Q
 
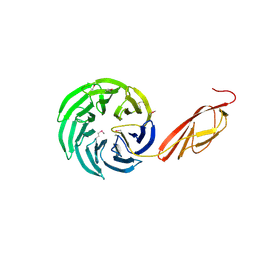 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
1K3R
 
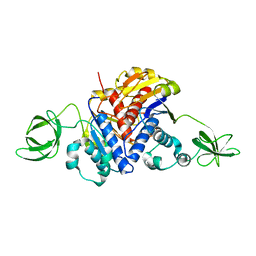 | | Crystal Structure of the Methyltransferase with a Knot from Methanobacterium thermoautotrophicum | | Descriptor: | conserved protein MT0001 | | Authors: | Zarembinski, T.I, Kim, Y, Peterson, K, Christendat, D, Dharamsi, A, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-03 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deep trefoil knot implicated in RNA binding found in an archaebacterial protein.
Proteins, 50, 2003
|
|
1L8R
 
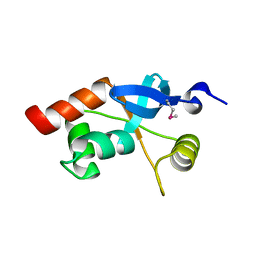 | | Structure of the Retinal Determination Protein Dachshund Reveals a DNA-Binding Motif | | Descriptor: | Dachshund | | Authors: | Kim, S.S, Zhang, R, Braunstein, S.E, Joachimiak, A, Cvekl, A, Hegde, R.S. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the retinal determination protein Dachshund reveals a DNA binding motif.
Structure, 10, 2002
|
|
1MK4
 
 | | Structure of Protein of Unknown Function YqjY from Bacillus subtilis, Probable Acetyltransferase | | Descriptor: | Hypothetical protein yqjY | | Authors: | Zhang, R, Dementiva, I, Mo, A, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-28 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7A crystal structure of a hypothetical protein yqjY
from Bacillus subtilis
To be Published
|
|
3C5P
 
 | | Crystal structure of BAS0735, a protein of unknown function from Bacillus anthracis str. Sterne | | Descriptor: | MAGNESIUM ION, Protein BAS0735 of unknown function | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-01 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of BAS0735, a protein of unknown function from Bacillus anthracis str. Sterne.
To be Published
|
|
4INJ
 
 | | Crystal structure of the S111A mutant of member of MccF clade from Listeria monocytogenes EGD-e with product | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lmo1638 protein, ... | | Authors: | Nocek, B, Tikhonov, A, Severinov, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the S111A mutant of member of MccF clade from Listeria monocytogenes EGD-e with product
To be Published
|
|
4RI0
 
 | | Serine Protease HtrA3, mutationally inactivated | | Descriptor: | PHOSPHATE ION, Serine protease HTRA3 | | Authors: | Osipiuk, J, Glaza, P, Wenta, T, Lipinska, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.272 Å) | | Cite: | Structural and Functional Analysis of Human HtrA3 Protease and Its Subdomains.
Plos One, 10, 2015
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
4KSN
 
 | |
1W8I
 
 | | The Structure of gene product af1683 from Archaeoglobus fulgidus. | | Descriptor: | PUTATIVE VAPC RIBONUCLEASE AF_1683 | | Authors: | Midwest Center for Structural Genomics (MCSG), Cuff, M.E, Zhang, R, Ginell, S.L, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Gene Product Af1683 from Archaeoglobus Fulgidus
To be Published
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4K08
 
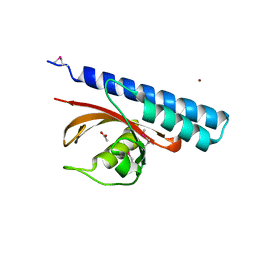 | | Periplasmic sensor domain of chemotaxis protein, Adeh_3718 | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis sensory transducer, ZINC ION | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-07-17 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1YTL
 
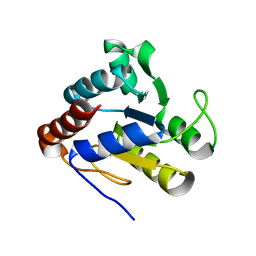 | | Crystal Structure of Acetyl-CoA decarboxylase/synthase complex epsilon subunit 2 | | Descriptor: | Acetyl-CoA decarboxylase/synthase complex epsilon subunit 2 | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-10 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Acetyl-CoA decarboxylase/synthase complex epsilon subunit 2 from Archaeoglobus fulgidus
To be Published
|
|
1YOY
 
 | |
1Y0N
 
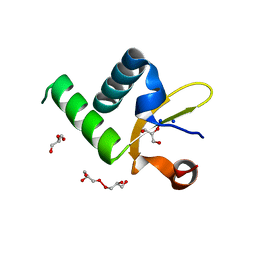 | | Structure of Protein of Unknown Function PA3463 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Hypothetical UPF0270 protein PA3463 | | Authors: | Binkowski, T.A, Edwards, A, Savchenko, A, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical protein PA3463 from Pseudomonas aeruginosa strain PAO1
To be Published
|
|
4JVO
 
 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with alanyl sulfamoyl adenylates | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, GLYCEROL, Microcin immunity protein MccF | | Authors: | Nocek, B, Tikhonov, A, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-25 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with alanyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
