4UW3
 
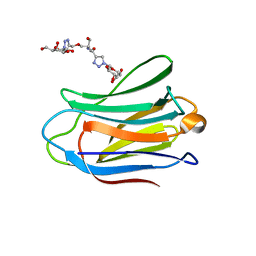 | | Human galectin-7 in complex with a galactose based dendron D1. | | Descriptor: | DENDRON-D1, HUMAN GALECTIN-7 | | Authors: | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
6ERH
 
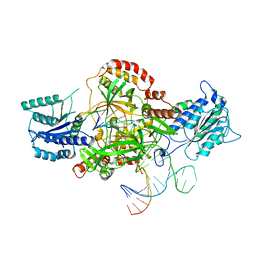 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6E6C
 
 | | HRAS G13D bound to GppNHp (H13GNP) | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
8RU1
 
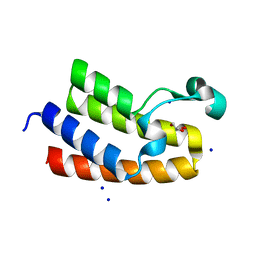 | | Chromatin remodeling regulator CECR2 with in crystallo disulfide bond | | Descriptor: | Chromatin remodeling regulator CECR2, GLYCEROL, SODIUM ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-01-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
4V39
 
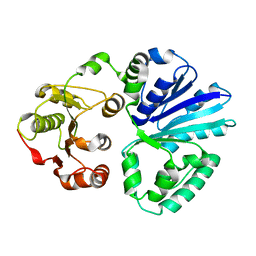 | | Apo-structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
8RU5
 
 | | ATPase family AAA domain containing 2 with crystallization epitope mutations V1022R:Q1027E | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2 | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-01-30 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
4V04
 
 | | FGFR1 in complex with ponatinib. | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4UU2
 
 | | Ferulic acid decarboxylase from Enterobacter sp., single mutant | | Descriptor: | FERULIC ACID DECARBOXYLASE, GLYCINE, PHOSPHATE ION, ... | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
4UYJ
 
 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
4UXH
 
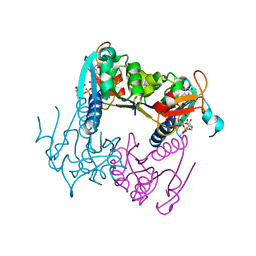 | | Leishmania major Thymidine Kinase in complex with AP5dT | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, THYMIDINE KINASE, ZINC ION | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
4UQU
 
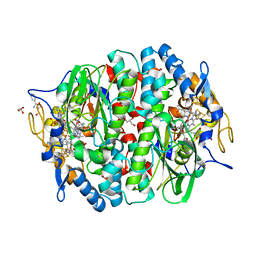 | | Crystal structure of the tetrachloroethene reductive dehalogenase from Sulfurospirillum multivorans | | Descriptor: | BENZAMIDINE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
4UXQ
 
 | | FGFR4 in complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, FIBROBLAST GROWTH FACTOR RECEPTOR 4, SULFATE ION | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4V1X
 
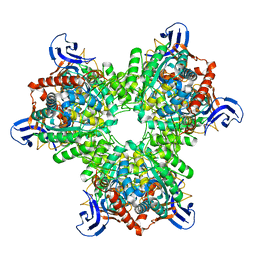 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4V5K
 
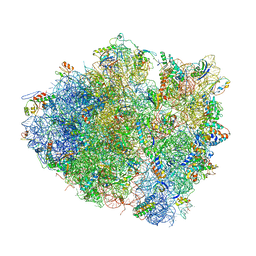 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4V70
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate pre-translocation state (pre3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6ERF
 
 | | Complex of APLF factor and Ku heterodimer bound to DNA | | Descriptor: | Aprataxin and PNK-like factor, DNA (34-MER), DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*GP*GP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat.Struct.Mol.Biol., 25, 2018
|
|
4V7E
 
 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | Authors: | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
4UTU
 
 | | Structural and biochemical characterization of the N- acetylmannosamine-6-phosphate 2-epimerase from Clostridium perfringens | | Descriptor: | CHLORIDE ION, N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE, N-acetylmannosamine-6-phosphate | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4UWM
 
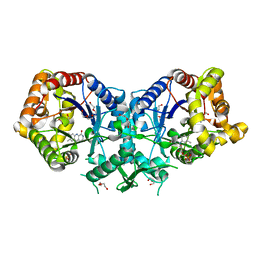 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WFS
 
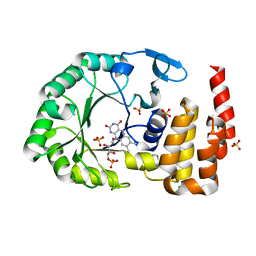 | | Crystal Structure of tRNA-dihydrouridine(20) synthase catalytic domain | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
6EXM
 
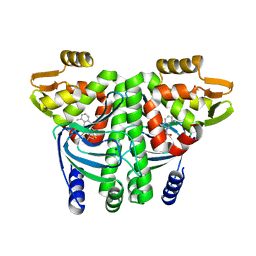 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK202) | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Grundstrom, C, Begum, A, Hall, M, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EYY
 
 | |
4WHE
 
 | | Crystal structure of E. coli phage shock protein A (PspA 1-144) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phage shock protein A | | Authors: | Parthier, C, Schoepfel, M, Stubbs, M.T, Osadnik, H, Brueser, T. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PspF-binding domain PspA1-144 and the PspAF complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins.
Mol.Microbiol., 98, 2015
|
|
6F6O
 
 | |
6EUT
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK67) | | Descriptor: | (3~{R})-8-cyclopropyl-7-[(4-methylnaphthalen-1-yl)methyl]-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
