1C78
 
 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
8I3X
 
 | | Rice APIP6-RING homodimer | | Descriptor: | RING-type domain-containing protein, ZINC ION | | Authors: | Zheng, Y, Zhang, X, Liu, Y, Liu, J, Wang, D. | | Deposit date: | 2023-01-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of rice APIP6 reveals a new dimerization mode of RING-type E3 ligases that facilities the construction of its working model
Phytopathol Res, 5, 2023
|
|
2KA9
 
 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | Descriptor: | Disks large homolog 4, cypin peptide | | Authors: | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
8I4F
 
 | | Omicron spike variant XBB with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4G
 
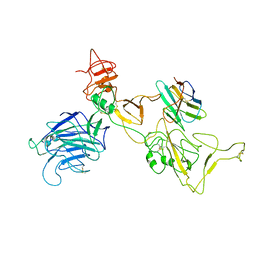 | | Omicron spike variant BQ.1.1 with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4E
 
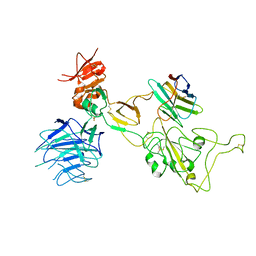 | | Omicron spike variant XBB with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4H
 
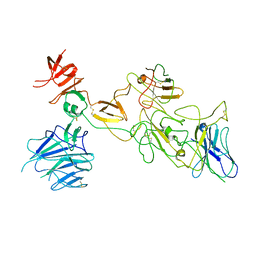 | | Omicron spike variant BA.1 with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
9JEA
 
 | |
9IK4
 
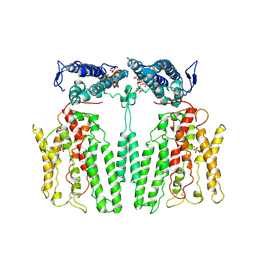 | |
7QTQ
 
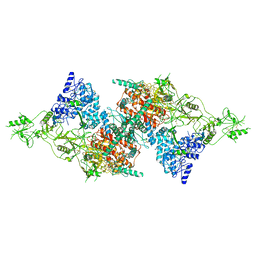 | | Structure of Native, iodinated bovine thyroglobulin solved on strepavidin affinity grids. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thyroglobulin, ... | | Authors: | Marechal, N, Weitz, J.C, Serrano, B.P, Zhang, X. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Formation of thyroid hormone revealed by a cryo-EM structure of native bovine thyroglobulin.
Nat Commun, 13, 2022
|
|
9JF8
 
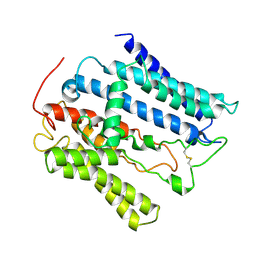 | |
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
5YC6
 
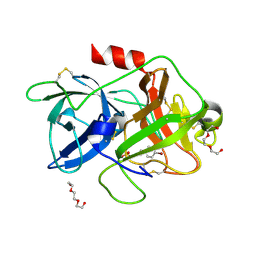 | | The crystal structure of uPA in complex with 4-Bromobenzylamirne at pH4.6 | | Descriptor: | 1-(4-BROMOPHENYL)METHANAMINE, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Zhang, X, Huang, M.D. | | Deposit date: | 2017-09-06 | | Release date: | 2018-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 8, 2018
|
|
5YCO
 
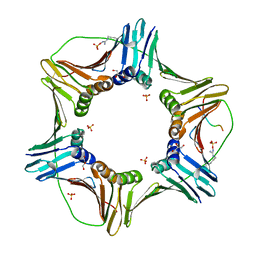 | | Complex structure of PCNA with UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
5YC7
 
 | |
4AM6
 
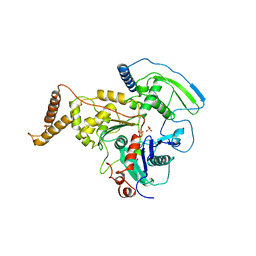 | | C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, SULFATE ION | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AM7
 
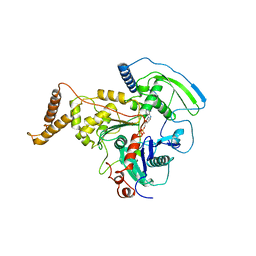 | | ADP-BOUND C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AU6
 
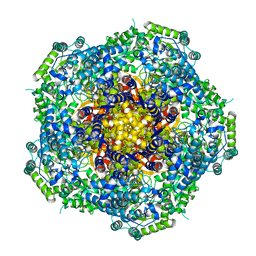 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
8WCN
 
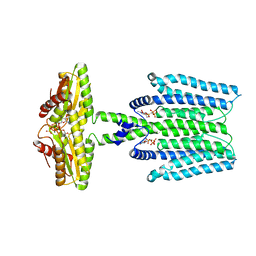 | | Cryo-EM structure of PAO1-ImcA with GMPCPP | | Descriptor: | Diguanylate cyclase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Zhan, X.L, Zhang, K, Wang, C.C, Fan, Q, Tang, X.J, Zhang, X, Wang, K, Fu, Y, Liang, H.H. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A c-di-GMP signaling module controls responses to iron in Pseudomonas aeruginosa.
Nat Commun, 15, 2024
|
|
5Z1C
 
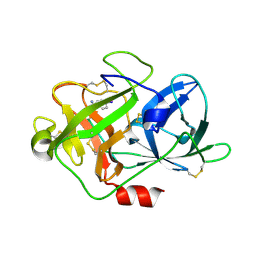 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
7SII
 
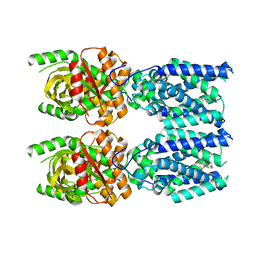 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | Authors: | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
4C6T
 
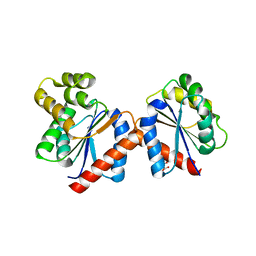 | | Crystal structure of the RPS4 and RRS1 TIR domain heterodimer | | Descriptor: | DISEASE RESISTANCE PROTEIN RPS4, MALONIC ACID, PROBABLE WRKY TRANSCRIPTION FACTOR 52 | | Authors: | Williams, S.J, Sohn, K.H, Wan, L, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
5WWL
 
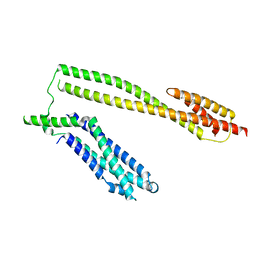 | | Crystal structure of the Schizogenesis pombe kinetochore Mis12C subcomplex | | Descriptor: | Centromere protein mis12, Kinetochore protein nnf1 | | Authors: | Wang, C, Zhou, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2017-01-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4C6R
 
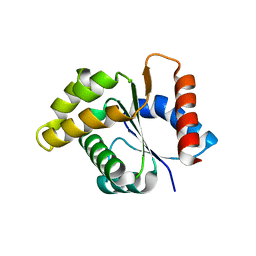 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPS4 | | Descriptor: | DISEASE RESISTANCE PROTEIN RPS4 | | Authors: | Williams, S.J, Sohn, K.H, Wan, L, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
4C6S
 
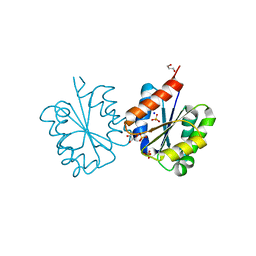 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RRS1 | | Descriptor: | GLYCEROL, PROBABLE WRKY TRANSCRIPTION FACTOR 52, SODIUM ION, ... | | Authors: | Wan, L, Williams, S.J, Sohn, K.H, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
