6KTC
 
 | | Crystal structure of YBX1 CSD with m5C RNA | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*GP*(5MC)P*CP*U)-3') | | Authors: | Zou, F, Li, S. | | Deposit date: | 2019-08-27 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | DrosophilaYBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
2YU2
 
 | | Crystal structure of hJHDM1A without a-ketoglutarate | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1A | | Authors: | Han, Z. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for histone demethylation by JHDM1
To be Published
|
|
2YU1
 
 | | Crystal structure of hJHDM1A complexed with a-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1A | | Authors: | Han, Z. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for histone demethylation by JHDM1
To be Published
|
|
3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
3LGP
 
 | | Crystal structure of catalytic domain of tace with benzimidazolyl-thienyl-tartrate based inhibitor | | Descriptor: | (2R,3R)-4-[(2R)-2-(3-chlorophenyl)pyrrolidin-1-yl]-2,3-dihydroxy-4-oxo-N-[(5-{[2-(trifluoromethyl)-1H-benzimidazol-1-yl]methyl}thiophen-2-yl)methyl]butanamide, Disintegrin and metalloproteinase domain-containing protein 17, ZINC ION | | Authors: | Orth, P. | | Deposit date: | 2010-01-21 | | Release date: | 2010-07-28 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity relationships of tartrate-based TACE inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
1KBO
 
 | | Complex of Human recombinant NAD(P)H:Quinone Oxide reductase type 1 with 5-methoxy-1,2-dimethyl-3-(phenoxymethyl)indole-4,7-dione (ES1340) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(PHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
3O64
 
 | | Crystal structure of catalytic domain of TACE with 2-(2-Aminothiazol-4-yl)pyrrolidine-Based Tartrate Diamides | | Descriptor: | (2R,3R)-2,3-dihydroxy-4-{(2R)-2-[2-(methylamino)-5-(methylsulfonyl)-1,3-thiazol-4-yl]pyrrolidin-1-yl}-4-oxo-N-{(1R)-1-[4-(1H-pyrazol-1-yl)phenyl]ethyl}butanamide, CALCIUM ION, ISOPROPYL ALCOHOL, ... | | Authors: | Orth, P. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 2-(2-Aminothiazol-4-yl)pyrrolidine-based tartrate diamides as potent, selective and orally bioavailable TACE inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1KBQ
 
 | | Complex of Human NAD(P)H quinone Oxidoreductase with 5-methoxy-1,2-dimethyl-3-(4-nitrophenoxymethyl)indole-4,7-dione (ES936) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(4-NITROPHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
6IIU
 
 | | Crystal structure of the human thromboxane A2 receptor bound to ramatroban | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[(3R)-3-[(4-fluorophenyl)sulfonylamino]-1,2,3,4-tetrahydrocarbazol-9-yl]propanoic acid, CHOLESTEROL, ... | | Authors: | Fan, H, Zhao, Q, Wu, B. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for ligand recognition of the human thromboxane A2receptor.
Nat. Chem. Biol., 15, 2019
|
|
6IIV
 
 | | Crystal structure of the human thromboxane A2 receptor bound to daltroban | | Descriptor: | 2-[4-[2-[(4-chlorophenyl)sulfonylamino]ethyl]phenyl]ethanoic acid, CHOLESTEROL, GLYCEROL, ... | | Authors: | Fan, H, Zhao, Q, Wu, B. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for ligand recognition of the human thromboxane A2receptor.
Nat. Chem. Biol., 15, 2019
|
|
7WOO
 
 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOT
 
 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
4IOP
 
 | | Crystal structure of NKp65 bound to its ligand KACL | | Descriptor: | C-type lectin domain family 2 member A, Killer cell lectin-like receptor subfamily F member 2, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y. | | Deposit date: | 2013-01-08 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of NKp65 bound to its keratinocyte ligand reveals basis for genetically linked recognition in natural killer gene complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5BNV
 
 | |
5UCW
 
 | |
5CJX
 
 | |
2IOA
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and ADP and phosphinate inhibitor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, D-GAMMA-GLUTAMYL-N-{[(R)-{4-[(4-AMINOBUTYL)AMINO]BUTYL}(PHOSPHONOOXY)PHOSPHORYL]METHYL}-D-ALANINAMIDE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IO9
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ ,GSH and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
7DTZ
 
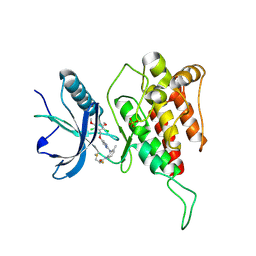 | | FGFR4 complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[5-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]methoxy]pyrimidin-2-yl]amino]-3-methyl-phenyl]-2-fluoranyl-prop-2-enamide, SULFATE ION | | Authors: | Chen, X.J, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-01-07 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Investigation of Covalent Warheads in the Design of 2-Aminopyrimidine-based FGFR4 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5DW3
 
 | |
5DVZ
 
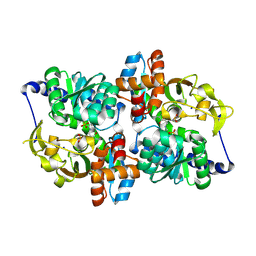 | | Holo TrpB from Pyrococcus furiosus | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Buller, A.R, Arnold, F.H. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-03 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Directed evolution of the tryptophan synthase beta-subunit for stand-alone function recapitulates allosteric activation.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5E0K
 
 | |
