4R12
 
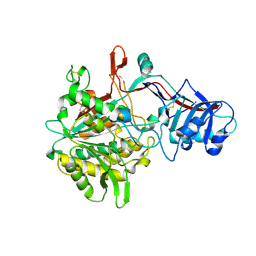 | | Crystal structure of the gamma-secretase component Nicastrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, T, Yan, C, Zhou, R, Zhao, Y, Sun, L, Yang, G, Lu, P, Ma, D, Shi, Y. | | Deposit date: | 2014-08-03 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the gamma-secretase component nicastrin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6TJO
 
 | | Cryo-EM structure of TypeI tau filaments extracted from the brains of individuals with Corticobasal degeneration | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Murzin, A.G, Falcon, B, Shi, Y, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-11-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Novel tau filament fold in corticobasal degeneration.
Nature, 580, 2020
|
|
6TJX
 
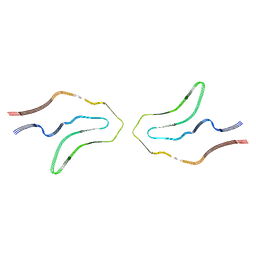 | | Cryo-EM structure of TypeII tau filaments extracted from the brains of individuals with Corticobasal degeneration | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Murzin, A.G, Falcon, B, Shi, Y, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Novel tau filament fold in corticobasal degeneration.
Nature, 580, 2020
|
|
3NSS
 
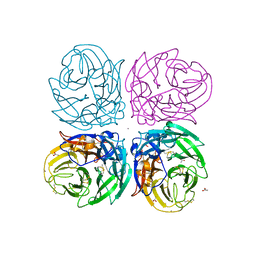 | | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active sites | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J.X, Zhang, W, Vavricka, C.J, Shi, Y, Gao, G.F. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site
Nat.Struct.Mol.Biol., 17, 2010
|
|
3P5N
 
 | |
5B88
 
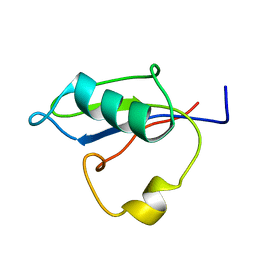 | | RRM-like domain of DEAD-box protein, CsdA | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Peng, J, Zhang, J, Wu, J, Tang, Y, Shi, Y. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5IY3
 
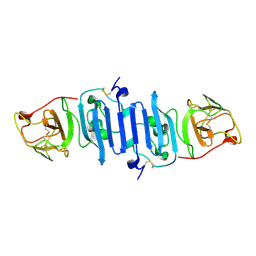 | | Zika Virus Non-structural Protein NS1 | | Descriptor: | Genome polyprotein | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses
Nat.Struct.Mol.Biol., 23, 2016
|
|
5CJB
 
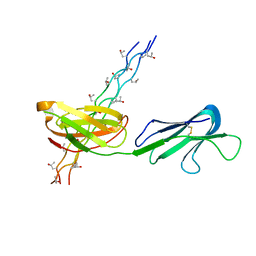 | | Human Osteoclast Associated Receptor (OSCAR) in complex with a collagen-like peptide | | Descriptor: | Osteoclast-associated immunoglobulin-like receptor, collagen-like peptide | | Authors: | Gao, G.F, Qi, J, Shi, Y, Haywood, J. | | Deposit date: | 2015-07-14 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of collagen recognition by human osteoclast-associated receptor and design of osteoclastogenesis inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JNX
 
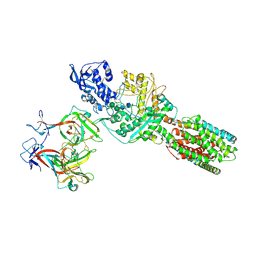 | | The 6.6 A cryo-EM structure of the full-length human NPC1 in complex with the cleaved glycoprotein of Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Wan, T, Shi, Y, Gao, F, Zhou, Q, Yan, N. | | Deposit date: | 2016-05-01 | | Release date: | 2016-06-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (6.56 Å) | | Cite: | Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection
Cell, 165, 2016
|
|
4DNV
 
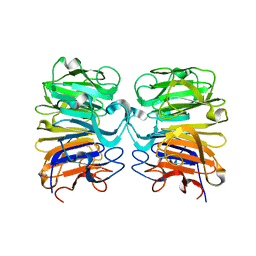 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNW
 
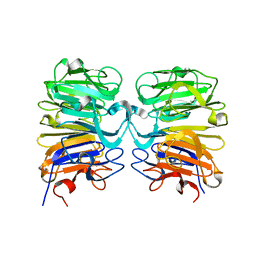 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
5CJ8
 
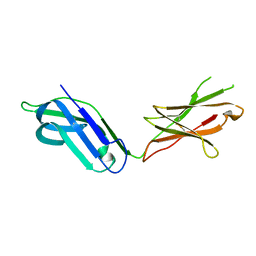 | |
4F23
 
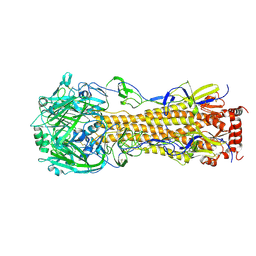 | | Influenza A virus hemagglutinin H16 HA0 structure with an alpha-helix conformation in the cleavage site: a potential drug target | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-07 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
4FIU
 
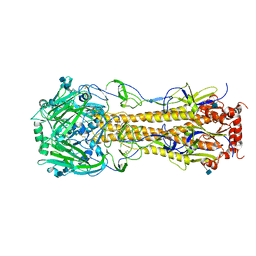 | | The structure of hemagglutinin of H16 subtype influenza virus with V327G mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Wang, M, Qi, J, Gao, G.F. | | Deposit date: | 2012-06-11 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
1FEW
 
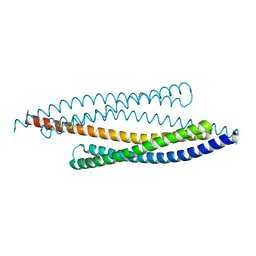 | | CRYSTAL STRUCTURE OF SMAC/DIABLO | | Descriptor: | SECOND MITOCHONDRIA-DERIVED ACTIVATOR OF CASPASES | | Authors: | Chai, J, Shi, Y. | | Deposit date: | 2000-07-23 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical basis of apoptotic activation by Smac/DIABLO.
Nature, 406, 2000
|
|
1G73
 
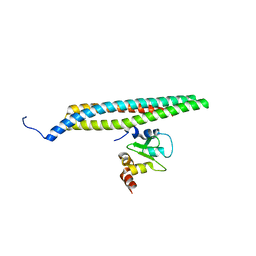 | | CRYSTAL STRUCTURE OF SMAC BOUND TO XIAP-BIR3 DOMAIN | | Descriptor: | INHIBITORS OF APOPTOSIS-LIKE PROTEIN ILP, SECOND MITOCHONDRIA-DERIVED ACTIVATOR OF CASPASES, ZINC ION | | Authors: | Wu, G, Chai, J, Suber, T.L, Wu, J.-W, Shi, Y. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of IAP recognition by Smac/DIABLO.
Nature, 408, 2000
|
|
1F0K
 
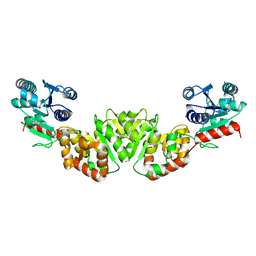 | | THE 1.9 ANGSTROM CRYSTAL STRUCTURE OF E. COLI MURG | | Descriptor: | SULFATE ION, UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE | | Authors: | Ha, S, Walker, D, Shi, Y, Walker, S. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis.
Protein Sci., 9, 2000
|
|
1HW5
 
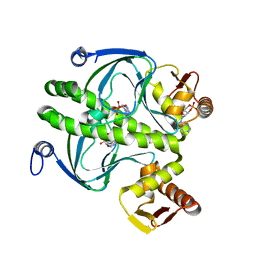 | | THE CAP/CRP VARIANT T127L/S128A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR | | Authors: | Chu, S.Y, Tordova, M, Gilliland, G.L, Gorshkova, I, Shi, Y. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding
J.Biol.Chem., 276, 2001
|
|
3F4M
 
 | | Crystal structure of TIPE2 | | Descriptor: | CHLORIDE ION, Tumor necrosis factor, alpha-induced protein 8-like protein 2 | | Authors: | Zhang, X, Wang, J, Shi, Y. | | Deposit date: | 2008-11-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of TIPE2 provides insights into immune homeostasis
Nat.Struct.Mol.Biol., 16, 2009
|
|
4YDE
 
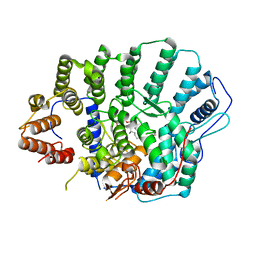 | | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE BINARY COMPLEX WITH THE ISOPRENOID FARNESYLDIPHOSPHATE | | Descriptor: | (3R,7S)-3,7,11-trimethyldodecyl trihydrogen diphosphate, 1,2-ETHANEDIOL, Protein farnesyltransferase/geranylgeranyltransferase type-1 Subunit beta, ... | | Authors: | Kumar, S, Mabanglo, M.F, Hast, M.A, Shi, Y, Beese, L.S. | | Deposit date: | 2015-02-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE BINARY COMPLEX WITH THE ISOPRENOID FARNESYLDIPHOSPHATE
To Be Published
|
|
4Y6K
 
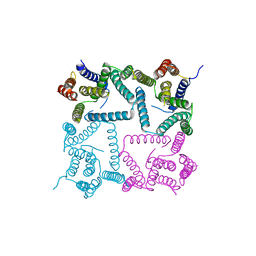 | | Complex structure of presenilin homologue PSH bound to an inhibitor | | Descriptor: | N-{(2R,4S,5S)-2-benzyl-5-[(tert-butoxycarbonyl)amino]-4-hydroxy-6-phenylhexanoyl}-L-leucyl-L-phenylalaninamide, Uncharacterized protein PSH | | Authors: | Dang, S, Wu, S, Wang, J, Shi, Y. | | Deposit date: | 2015-02-13 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.855 Å) | | Cite: | Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3H13
 
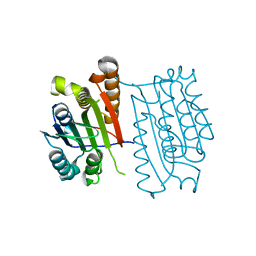 | | c-FLIPL protease-like domain | | Descriptor: | CASP8 and FADD-like apoptosis regulator | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of procaspase-8 activation by c-FLIPL.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H4M
 
 | | AAA ATPase domain of the proteasome- activating nucleotidase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | Authors: | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H43
 
 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | Descriptor: | Proteasome-activating nucleotidase | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
