1IQD
 
 | | Human Factor VIII C2 Domain complexed to human monoclonal BO2C11 Fab. | | Descriptor: | HUMAN FACTOR VIII, HUMAN MONOCLONAL BO2C11 FAB HEAVY CHAIN, HUMAN MONOCLONAL BO2C11 FAB LIGHT CHAIN | | Authors: | Spiegel Jr, P.C, Jacquemin, M, Saint-Remy, J.M, Stoddard, B.L, Pratt, K.P. | | Deposit date: | 2001-07-21 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a factor VIII C2 domain-immunoglobulin G4kappa Fab complex: identification of an inhibitory antibody epitope on the surface of factor VIII.
Blood, 98, 2001
|
|
6XR1
 
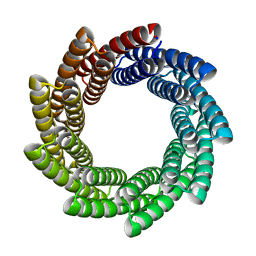 | |
6XR2
 
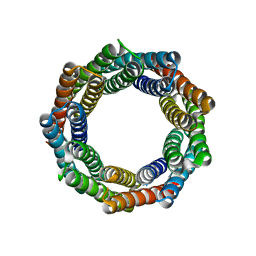 | |
6CZG
 
 | |
6CZJ
 
 | |
6CZH
 
 | | Structure of a redesigned beta barrel, mFAP0, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP0 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
7UNJ
 
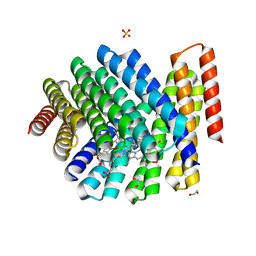 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
2ERH
 
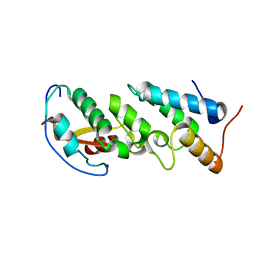 | | Crystal Structure of the E7_G/Im7_G complex; a designed interface between the colicin E7 DNAse and the Im7 immunity protein | | Descriptor: | Colicin E7, Colicin E7 immunity protein | | Authors: | Joachimiak, L.A, Kortemme, T, Stoddard, B.L, Baker, D. | | Deposit date: | 2005-10-24 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of a New Hydrogen Bond Network and at Least a 300-fold Specificity Switch at a Protein-Protein Interface.
J.Mol.Biol., 361, 2006
|
|
2EX5
 
 | |
2FLD
 
 | | I-MsoI Re-Designed for Altered DNA Cleavage Specificity | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Ashworth, J, Duarte, C.M, Havranek, J.J, Sussman, D, Monnat, R.J, Stoddard, B.L, Baker, D. | | Deposit date: | 2006-01-05 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational redesign of endonuclease DNA binding and cleavage specificity.
Nature, 441, 2006
|
|
6UVW
 
 | |
6UWK
 
 | |
6UWJ
 
 | |
6UWH
 
 | |
6UWG
 
 | |
6UW0
 
 | |
7LVV
 
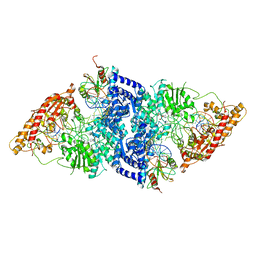 | | cryoEM structure DrdV-DNA complex | | Descriptor: | CALCIUM ION, DNA (27-MER), DNA (28-MER), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.
Structure, 29, 2021
|
|
7LO5
 
 | | cryoEM structure DrdV-DNA complex | | Descriptor: | CALCIUM ION, DNA (27-MER), DNA (28-MER), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2021-02-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.
Structure, 29, 2021
|
|
1IPP
 
 | | HOMING ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED ENDONUCLEASE I-PPOI, ... | | Authors: | Flick, K.E, Jurica, M.S, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
1DT0
 
 | | CLONING, SEQUENCE, AND CRYSTALLOGRAPHIC STRUCTURE OF RECOMBINANT IRON SUPEROXIDE DISMUTASE FROM PSEUDOMONAS OVALIS | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bond, C.J, Huang, J, Hajduk, R, Flick, K, Heath, P, Stoddard, B.L. | | Deposit date: | 2000-01-10 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cloning, sequence and crystallographic structure of recombinant iron superoxide dismutase from Pseudomonas ovalis.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EVW
 
 | | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*TP*AP*CP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
1EVX
 
 | | APO CRYSTAL STRUCTURE OF THE HOMING ENDONUCLEASE, I-PPOI | | Descriptor: | INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, SULFATE ION, ZINC ION | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
