1SCW
 
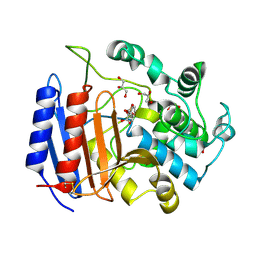 | | TOWARD BETTER ANTIBIOTICS: CRYSTAL STRUCTURE OF R61 DD-PEPTIDASE INHIBITED BY A NOVEL MONOCYCLIC PHOSPHATE INHIBITOR | | Descriptor: | (2Z)-3-{[OXIDO(OXO)PHOSPHINO]OXY}-2-PHENYLACRYLATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Kaur, K, Adediran, S.A, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Toward Better Antibiotics: Crystallographic Studies of a Novel Class of DD-Peptidase/beta-Lactamase Inhibitors.
Biochemistry, 43, 2004
|
|
1RAT
 
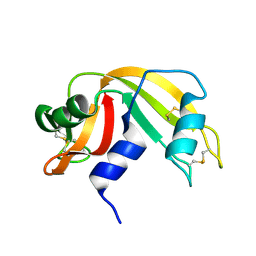 | |
1KQ1
 
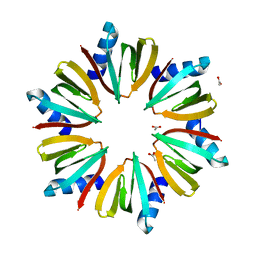 | | 1.55 A Crystal structure of the pleiotropic translational regulator, Hfq | | Descriptor: | ACETIC ACID, Host Factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
1KQ2
 
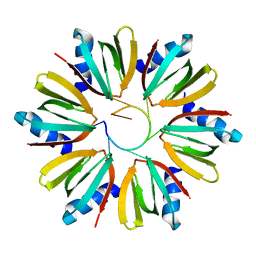 | | Crystal Structure of an Hfq-RNA Complex | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*G)-3', Host factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
5LYT
 
 | |
4V6N
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (50S ribosome of class2 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6L
 
 | | Structural insights into cognate vs. near-cognate discrimination during decoding. | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Agirrezabala, X, Schreiner, E, Trabuco, L.G, Lei, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-01-07 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (13.2 Å) | | Cite: | Structural insights into cognate versus near-cognate discrimination during decoding.
Embo J., 30, 2011
|
|
4V6K
 
 | | Structural insights into cognate vs. near-cognate discrimination during decoding. | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Agirrezabala, X, Schreiner, E, Trabuco, L.G, Lei, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-01-07 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (8.25 Å) | | Cite: | Structural insights into cognate versus near-cognate discrimination during decoding.
Embo J., 30, 2011
|
|
4YND
 
 | | The Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2 | | Descriptor: | N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[(2R)-2-hydroxy-2-(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Sweis, R.F, Wang, Z, Algire, M, Arrowsmith, C.H, Brown, P.J, Chiang, G.C, Guo, J, Jakob, C.G, Kennedy, S, Li, F, Soni, N.B, Vedadi, M, Pappano, W.N. | | Deposit date: | 2015-03-09 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2.
Acs Med.Chem.Lett., 6, 2015
|
|
1N8E
 
 | | Fragment Double-D from Human Fibrin | | Descriptor: | Fibrin alpha/alpha-E chain, Fibrin beta chain, Fibrin gamma chain | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The Crystal Structure of Fragment Double-D from Cross-Linked Lamprey Fibrin Reveals Isopeptide Linkages across an Unexpected D-D Interface
Biochemistry, 41, 2002
|
|
1MPL
 
 | | CRYSTAL STRUCTURE OF PHOSPHONATE-INHIBITED D-ALA-D-ALA PEPTIDASE REVEALS AN ANALOG OF A TETRAHEDRAL TRANSITION STATE | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, GLYCYL-L-A-AMINOPIMELYL-E-(D-2-AMINOETHYL)PHOSPHONATE | | Authors: | Silvaggi, N.R, Anderson, J.W, Brinsmade, S.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2002-09-12 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Crystal Structure of Phosphonate-Inhibited
d-Ala-d-Ala Peptidase Reveals an Analogue of a Tetrahedral Transition State.
Biochemistry, 42, 2003
|
|
3RAT
 
 | |
6LYT
 
 | |
449D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
445D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', Benzimidazole derivative complex | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
448D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODO-4-METHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Clark, G.R, Squire, C.J, Martin, R.F, White, J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
444D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODO-4-METHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Clark, G.R, Squire, C.J, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
442D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODO-4-METHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
1YXE
 
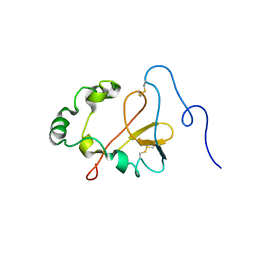 | | Structure and inter-domain interactions of domain II from the blood stage malarial protein, apical membrane antigen 1 | | Descriptor: | apical membrane antigen 1 | | Authors: | Feng, Z.P, Keizer, D.W, Stevenson, R.A, Yao, S, Babon, J.J, Murphy, V.J, Anders, R.F, Norton, R.S. | | Deposit date: | 2005-02-21 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and Inter-domain Interactions of Domain II from the Blood-stage Malarial Protein, Apical Membrane Antigen 1.
J.Mol.Biol., 350, 2005
|
|
6WAD
 
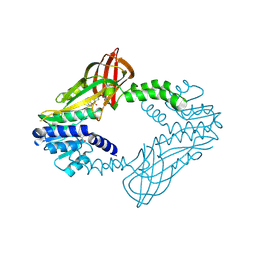 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with MT2739 inhibitor | | Descriptor: | 5-bromo-N-(diphenylmethyl)-N-methylthiophene-2-carboxamide, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Schapira, M, De Freitas, R.F, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with MT2739 inhibitor
to be published
|
|
443D
 
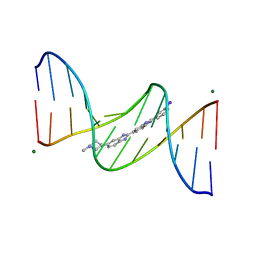 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'/ BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
4K68
 
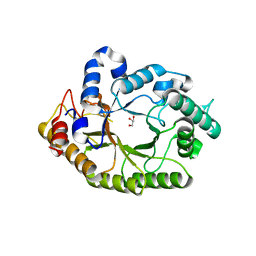 | | Structure of a novel GH10 endoxylanase retrieved from sugarcane soil metagenome | | Descriptor: | GH10 xylanase, GLYCEROL | | Authors: | Santos, C.R, Polo, C.C, Alvarez, T.M, Paixao, D.A.A, Almeida, R.F, Pereira, I.O, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Development and biotechnological application of a novel endoxylanase family GH10 identified from sugarcane soil metagenome.
Plos One, 8, 2013
|
|
447D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Descriptor: | 2'-(4-DIMETHYLAMINOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-18 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
1Z40
 
 | | AMA1 from Plasmodium falciparum | | Descriptor: | CHLORIDE ION, apical membrane antigen 1 precursor | | Authors: | Bai, T, Becker, M, Gupta, A, Strike, P, Murphy, V.J, Anders, R.F, Batchelor, A.H. | | Deposit date: | 2005-03-14 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZCM
 
 | | Human calpain protease core inhibited by ZLLYCH2F | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Li, Q, Hanzlik, R.P, Weaver, R.F, Schonbrunn, E. | | Deposit date: | 2005-04-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mode of action of a covalently inhibiting peptidomimetic on the human calpain protease core
Biochemistry, 45, 2006
|
|
