5NID
 
 | |
5NI6
 
 | | Crystal structure of human LTA4H mutant D375N in complex with LTA4 | | Descriptor: | 5S-5,6-oxido-7,9-trans-11,14-cis-eicosatetraenoic acid, ACETIC ACID, Leukotriene A-4 hydrolase, ... | | Authors: | Stsiapanava, A. | | Deposit date: | 2017-03-23 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Capturing LTA4 hydrolase in action: Insights to the chemistry and dynamics of chemotactic LTB4 synthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NI2
 
 | |
4DPR
 
 | | Structure of human Leukotriene A4 hydrolase in complex with inhibitor captopril | | Descriptor: | ACETIC ACID, GLYCEROL, L-CAPTOPRIL, ... | | Authors: | Stsiapanava, A, Haeggstrom, J.Z, Rinaldo-Matthis, A. | | Deposit date: | 2012-02-14 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Capturing LTA4hydrolase in action: Insights to the chemistry and dynamics of chemotactic LTB4synthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NI4
 
 | |
5NIE
 
 | |
7PFP
 
 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3FWH
 
 | | Structure of haloalkane dehalogenase mutant Dha15 (I135F/C176Y) from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Dohnalek, J, Lapkouski, M, Kuta Smatanova, I. | | Deposit date: | 2009-01-18 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3G9X
 
 | | Structure of haloalkane dehalogenase DhaA14 mutant I135F from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Lapkouski, M, Dohnalek, J, Kuta Smatanova, I. | | Deposit date: | 2009-02-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3FBW
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant C176Y | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Dohnalek, J, Stsiapanava, A, Gavira, J.A, Kuta Smatanova, I, Kuty, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4P7T
 
 | |
4HZG
 
 | | Structure of haloalkane dehalogenase DhaA from Rhodococcus rhodochrous | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Stsiapanava, A, Weiss, M.S, Mesters, J.R, Kuta Smatanova, I. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4FA6
 
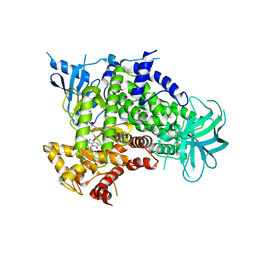 | |
7SIU
 
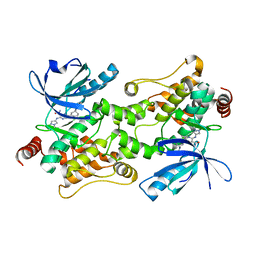 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
4FWB
 
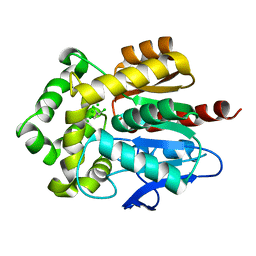 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 in complex with 1, 2, 3 - trichloropropane | | Descriptor: | 1,2,3-trichloropropane, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Kuta Smatanova, I. | | Deposit date: | 2012-06-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1CTM
 
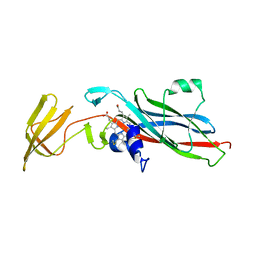 | | CRYSTAL STRUCTURE OF CHLOROPLAST CYTOCHROME F REVEALS A NOVEL CYTOCHROME FOLD AND UNEXPECTED HEME LIGATION | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1994-01-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation.
Structure, 2, 1994
|
|
7ZTZ
 
 | | Crystal structure of mutant AR-LBD (Y764C) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZTX
 
 | | Crystal structure of mutant AR-LBD (F755V) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZU2
 
 | | Crystal structure of mutant AR-LBD (Q799E) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZU1
 
 | | Crystal structure of mutant AR-LBD (V758A) bound to dihydrotestosterone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZTV
 
 | | Crystal structure of mutant AR-LBD (F755L) bound to dihydrotestosterone | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
8RKF
 
 | | Crystal structure of the ZP-N1 and ZP-N2 domains of human ZP2 (hZP2-N1N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Stsiapanava, A, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
3SK0
 
 | | structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant DhaA12 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Koudelakova, T, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2011-06-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dynamics and hydration explain failed functional transformation in dehalogenase design.
Nat.Chem.Biol., 10, 2014
|
|
3RJA
 
 | | Crystal structure of carbohydrate oxidase from Microdochium nivale in complex with substrate analogue | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duskova, J, Skalova, T, Kolenko, P, Stepankova, A, Koval, T, Hasek, J, Ostergaard, L.H, Fuglsang, C.C, Dohnalek, J. | | Deposit date: | 2011-04-15 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and kinetic studies of carbohydrate oxidase from Microdochium nivale
To be Published
|
|
