6UWV
 
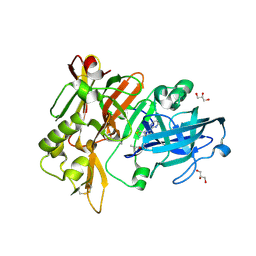 | | BACE-1 in complex with compound #34 | | Descriptor: | (4aR,7aR)-7a-[(1R,2R)-2-(2-{[(1R,2R)-2-methylcyclopropyl]methoxy}propan-2-yl)cyclopropyl]-6-(pyrimidin-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Stout, S.L. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVY
 
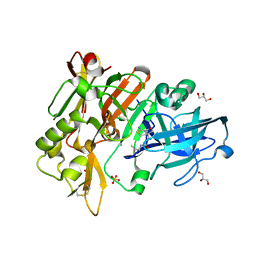 | | BACE-1 in complex with compound #18 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVV
 
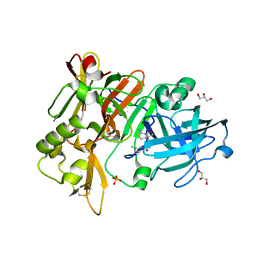 | | BACE-1 in complex with compound #17 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-butylcyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
3DKF
 
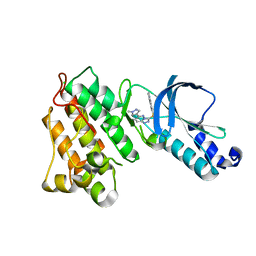 | | Structure of MET receptor tyrosine kinase in complex with inhibitor SGX-523 | | Descriptor: | 6-{[6-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}quinoline, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Hendle, J. | | Deposit date: | 2008-06-24 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SGX523 is an exquisitely selective, ATP-competitive inhibitor of the MET receptor tyrosine kinase with antitumor activity in vivo.
Mol.Cancer Ther., 8, 2009
|
|
3DKG
 
 | | Structure of Mutant(Y1248L) MET receptor tyrosine kinase in complex with inhibitor SGX-523 | | Descriptor: | 6-{[6-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}quinoline, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Hendle, J. | | Deposit date: | 2008-06-24 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SGX523 is an exquisitely selective, ATP-competitive inhibitor of the MET receptor tyrosine kinase with antitumor activity in vivo.
Mol.Cancer Ther., 8, 2009
|
|
3CIZ
 
 | |
3CJ5
 
 | |
3DKC
 
 | | Structure of MET receptor tyrosine kinase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Hendle, J. | | Deposit date: | 2008-06-24 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | SGX523 is an exquisitely selective, ATP-competitive inhibitor of the MET receptor tyrosine kinase with antitumor activity in vivo.
Mol.Cancer Ther., 8, 2009
|
|
2PZF
 
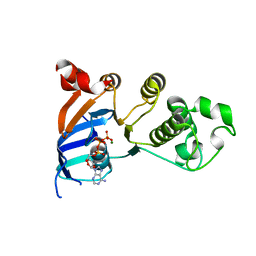 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer with delta F508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
1MDO
 
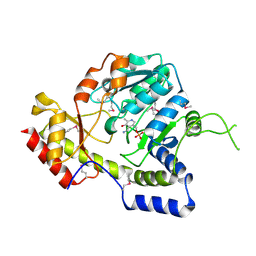 | | Crystal structure of ArnB aminotransferase with pyridomine 5' phosphate | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ArnB aminotransferase | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
2PZG
 
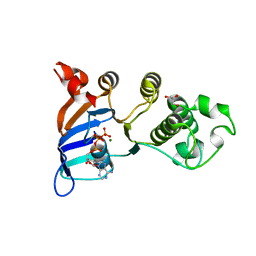 | | Minimal human CFTR first nucleotide binding domain as a monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
2PZE
 
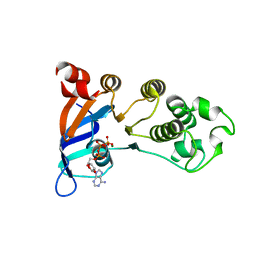 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
1MDZ
 
 | | Crystal structure of ArnB aminotransferase with cycloserine and pyridoxal 5' phosphate | | Descriptor: | ArnB aminotransferase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
1MDX
 
 | | Crystal structure of ArnB transferase with pyridoxal 5' phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ArnB aminotransferase, GLYCEROL, ... | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T.A, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
1GAL
 
 | | CRYSTAL STRUCTURE OF GLUCOSE OXIDASE FROM ASPERGILLUS NIGER: REFINED AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, GLUCOSE OXIDASE, ... | | Authors: | Hecht, H.J, Kalisz, K, Hendle, J, Schmid, R.D, Schomburg, D. | | Deposit date: | 1992-08-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glucose oxidase from Aspergillus niger refined at 2.3 A resolution.
J.Mol.Biol., 229, 1993
|
|
1Q2W
 
 | | X-Ray Crystal Structure of the SARS Coronavirus Main Protease | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like protease | | Authors: | Bonanno, J.B, Fowler, R, Gupta, S, Hendle, J, Lorimer, D, Romero, R, Sauder, J.M, Wei, C.L, Liu, E.T, Burley, S.K, Harris, T. | | Deposit date: | 2003-07-26 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Company Says It Mapped Part of SARS Virus
New York Times, 30 July, 2003
|
|
1XBA
 
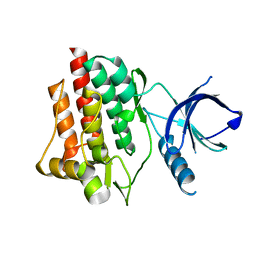 | | Crystal structure of apo syk tyrosine kinase domain | | Descriptor: | Tyrosine-protein kinase SYK | | Authors: | Atwell, S, Adams, J.M, Badger, J, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Nienaber, V.L, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
1XBB
 
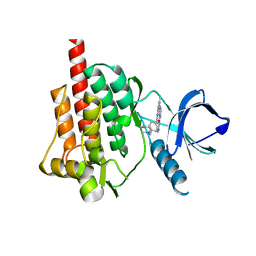 | | Crystal structure of the syk tyrosine kinase domain with Gleevec | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Tyrosine-protein kinase SYK | | Authors: | Nienaber, V.L, Atwell, S, Adams, J.M, Badger, J, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A Novel Mode of Gleevec Binding Is Revealed by the Structure of Spleen Tyrosine Kinase
J.Biol.Chem., 279, 2004
|
|
1XBC
 
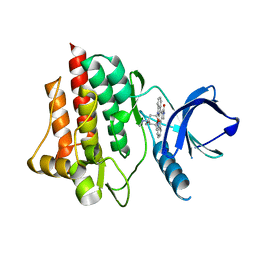 | | Crystal structure of the syk tyrosine kinase domain with Staurosporin | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase SYK | | Authors: | Badger, J, Atwell, S, Adams, J.M, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Nienaber, V.L, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase
J.Biol.Chem., 279, 2004
|
|
1CF3
 
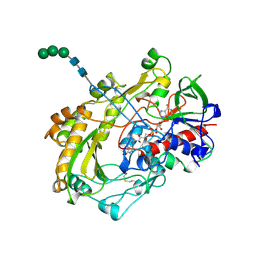 | | GLUCOSE OXIDASE FROM APERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (GLUCOSE OXIDASE), ... | | Authors: | Hecht, H.J, Kalisz, H. | | Deposit date: | 1999-03-23 | | Release date: | 1999-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6WGJ
 
 | | Fab portion of dupilumab with Crystal Kappa design and no interchain disulfide | | Descriptor: | Dupilumab Fab heavy chain, Dupilumab Fab light chain | | Authors: | Druzina, Z, Atwell, S, Pustilnik, A, Antonysamy, S, Ho, C, Lieu, R, Hendle, J, Benach, J, Wang, J. | | Deposit date: | 2020-04-05 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
7L3N
 
 | | SARS-CoV 2 Spike Protein bound to LY-CoV555 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV555 Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection.
Biorxiv, 2020
|
|
1VHN
 
 | |
1VI7
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yigZ | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YIGZ, a conserved hypothetical protein from Escherichia coli k12 with a novel fold
Proteins, 55, 2004
|
|
3QVP
 
 | | Crystal structure of glucose oxidase for space group C2221 at 1.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
