7TN0
 
 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
7L6W
 
 | | SFX structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Flueckiger, L, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Stacey, K.J, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
4ZXB
 
 | | Structure of the human insulin receptor ectodomain, IRDeltabeta construct, in complex with four Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Croll, T, Smith, B.J, Margetts, M.B, Whittaker, J, Weiss, M.A, Ward, C.W, Lawrence, M.C. | | Deposit date: | 2015-05-20 | | Release date: | 2016-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-Resolution Structure of the Human Insulin Receptor Ectodomain: Multi-Modal Inclusion of the Insert Domain.
Structure, 24, 2016
|
|
5KSD
 
 | | Crystal Structure of a Plasma Membrane Proton Pump | | Descriptor: | ATPase 2, plasma membrane-type, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Croll, T, Pedersen, B.P, Nissen, P. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Improved Model of Proton Pump Crystal Structure Obtained by Interactive Molecular Dynamics Flexible Fitting Expands the Mechanistic Model for Proton Translocation in P-Type ATPases.
Front Physiol, 8, 2017
|
|
6I2M
 
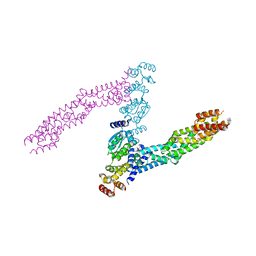 | |
5TEB
 
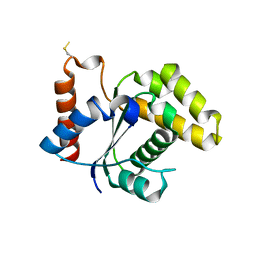 | | Crystal Structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPP1 | | Descriptor: | Recognition of Peronospora parasitica 1 | | Authors: | Bentham, A.R, Zhang, X, Croll, T, Williams, S, Kobe, B. | | Deposit date: | 2016-09-20 | | Release date: | 2017-02-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6OGD
 
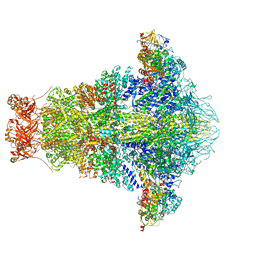 | | Cryo-EM structure of YenTcA in its prepore state | | Descriptor: | Chitinase 2, Toxin subunit YenA1, Toxin subunit YenA2 | | Authors: | Piper, S.J, Brillault, L, Box, J.K, Landsberg, M.J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures of the pore-forming A subunit from the Yersinia entomophaga ABC toxin.
Nat Commun, 10, 2019
|
|
6ZHY
 
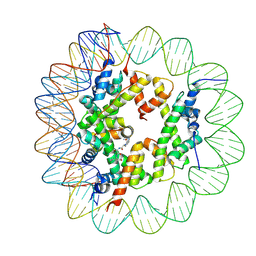 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
8EUO
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | to be published
To Be Published
|
|
7S1G
 
 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1H
 
 | | Wild-type Escherichia coli ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UPH
 
 | | Structure of a ribosome with tethered subunits | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kim, D.S, Watkins, A, Bidstrup, E, Lee, J, Topkar, V.V, Kofman, C, Schwarz, K.J, Liu, Y, Pintilie, G, Roney, E, Das, R, Jewett, M.C. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Three-dimensional structure-guided evolution of a ribosome with tethered subunits.
Nat.Chem.Biol., 18, 2022
|
|
7R7N
 
 | |
5TEC
 
 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | Descriptor: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | Authors: | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2016-09-20 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9EM1
 
 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone and phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, Chronophin, GLYCEROL, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
7O0W
 
 | | Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0U
 
 | | Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0X
 
 | | Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-28 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0V
 
 | | Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
8CKP
 
 | | X-ray structure of the crystallization-prone form of subfamily III haloalkane dehalogenase DhmeA from Haloferax mediterranei | | Descriptor: | Alpha/beta fold hydrolase, CHLORIDE ION | | Authors: | Marek, M, Chmelova, K, Schenkmayerova, A, Croll, T, Read, R.J, Diederichs, K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Multimeric structure of a subfamily III haloalkane dehalogenase-like enzyme solved by combination of cryo-EM and x-ray crystallography.
Protein Sci., 32, 2023
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8G8F
 
 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
7DW9
 
 | |
