3UIJ
 
 | |
3UIH
 
 | |
5WXV
 
 | |
7N29
 
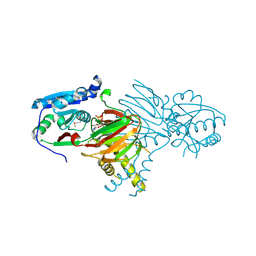 | | Structure of NAD kinase | | Descriptor: | NAD kinase 2, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Du, J, Estrella, M.A, Jeffrey, P.D, Korennykh, A.V. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human NADK2 reveals atypical assembly and regulation of NAD kinases from animal mitochondria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7EPC
 
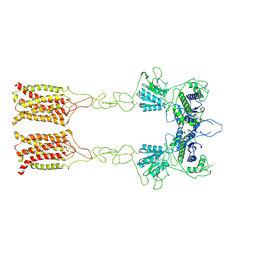 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | Descriptor: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPA
 
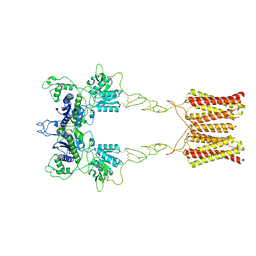 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPF
 
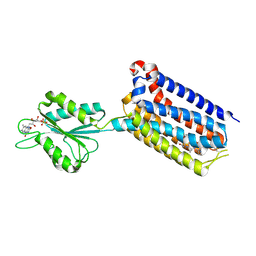 | | Crystal structure of mGlu2 bound to NAM597 | | Descriptor: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPB
 
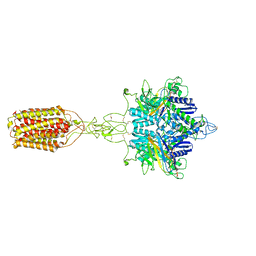 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
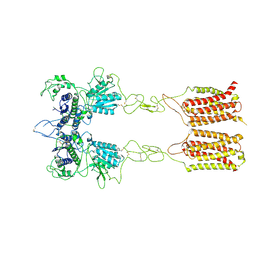 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
4ONJ
 
 | | Crystal structure of the catalytic domain of ntDRM | | Descriptor: | DNA methyltransferase, SINEFUNGIN | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular Mechanism of Action of Plant DRM De Novo DNA Methyltransferases.
Cell(Cambridge,Mass.), 157, 2014
|
|
4ONQ
 
 | |
6PUS
 
 | | Human TRPM2 bound to ADPR and calcium | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUR
 
 | | Human TRPM2 bound to ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUU
 
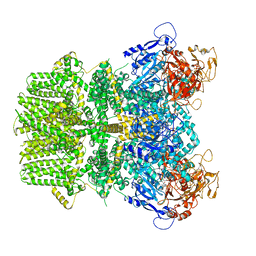 | | Human TRPM2 bound to 8-Br-cADPR and calcium | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUO
 
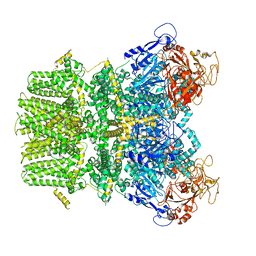 | | Human TRPM2 in the apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6DRJ
 
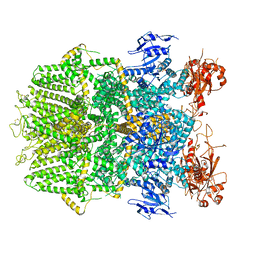 | | Structure of TRPM2 ion channel receptor by single particle electron cryo-microscopy, ADPR/Ca2+ bound state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, Transient receptor potential cation channel, ... | | Authors: | Du, J, Lu, W, Huang, Y, Winkler, P, Sun, W. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium.
Nature, 562, 2018
|
|
6YX5
 
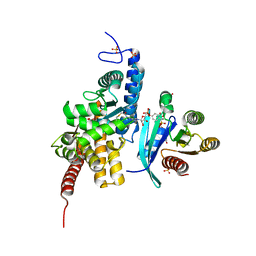 | | Structure of DrrA from Legionella pneumophilia in complex with human Rab8a | | Descriptor: | MAGNESIUM ION, Multifunctional virulence effector protein DrrA, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Schneider, S, Du, J, von Wrisberg, M.K, Lang, K, Itzen, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1.
Nat Commun, 12, 2021
|
|
8D8R
 
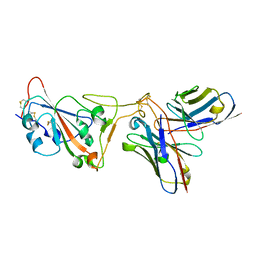 | | SARS-CoV-2 Spike RBD in complex with DMAb 2196 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2196 heavy chain, 2196 light chain, ... | | Authors: | Du, J, Cui, J, Pallesen, J. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Nat Commun, 13, 2022
|
|
8D8Q
 
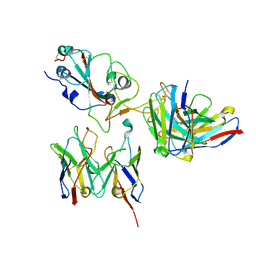 | | SARS-CoV-2 Spike RBD in complex with DMAbs 2130 and 2196 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2130 heavy chain, 2130 light chain, ... | | Authors: | Du, J, Cui, J, Pallesen, J. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Nat Commun, 13, 2022
|
|
6PXD
 
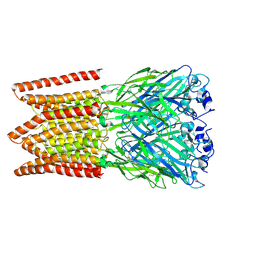 | | CryoEM structure of zebra fish alpha-1 glycine receptor, Apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Gouaux, E. | | Deposit date: | 2019-07-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6DRK
 
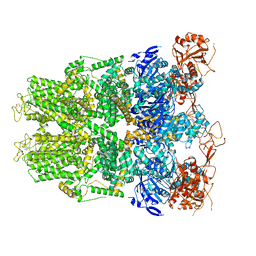 | | Structure of TRPM2 ion channel receptor by single particle electron cryo-microscopy, Apo state | | Descriptor: | Transient receptor potential cation channel, subfamily M, member 2 | | Authors: | Du, J, Lu, W, Huang, Y, Winkler, P, Sun, W. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium.
Nature, 562, 2018
|
|
8YHZ
 
 | | The co-crystal structure of the Fab fragment of Ab-1080 with NaV1.7 VSDII peptide | | Descriptor: | Heavy chain of 1080 Fab, Light chain of 1080 Fab, Sodium channel protein type 9 subunit alpha | | Authors: | Du, J, Zhang, Y, Zhu, R, Ding, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Intra-channel bi-epitopic crosslinking unleashes ultrapotent antibodies targeting Na V 1.7 for pain alleviation.
Cell Rep Med, 2024
|
|
4NJ5
 
 | | Crystal structure of SUVH9 | | Descriptor: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
4IUU
 
 | |
