6M6G
 
 | | Structure of HSV2 viron capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Coiled coils, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (5.39 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
8DCS
 
 | | Cryo-EM structure of cyanopindolol-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
7PKN
 
 | | Structure of the human CCAN deltaCT complex | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K, ... | | Authors: | Muir, K.W, Yatskevich, S, Bellini, D, Barford, D. | | Deposit date: | 2021-08-25 | | Release date: | 2022-04-27 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
8DCR
 
 | | Cryo-EM structure of dobutamine-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | DOBUTAMINE, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
5WRW
 
 | | Structure of human apo-SRP72 | | Descriptor: | SULFATE ION, Signal recognition particle subunit SRP72 | | Authors: | Gao, Y, Chen, Z. | | Deposit date: | 2016-12-04 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
J Mol Cell Biol, 9, 2017
|
|
7KHL
 
 | | BRD4-BD1 Compound6 (methyl 4-(3,5-difluoropyridin-2-yl)-10-methyl-7-((methylsulfonyl)methyl)-11-oxo-3,4,10,11-tetrahydro-1H-1,4,10-triazadibenzo[cd,f]azulene-6-carboxylate) | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, methyl 7-(3,5-difluoropyridin-2-yl)-2-methyl-10-[(methylsulfonyl)methyl]-3-oxo-3,4,6,7-tetrahydro-2H-2,4,7-triazadibenzo[cd,f]azulene-9-carboxylate | | Authors: | Murray, J.M. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.286 Å) | | Cite: | Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy.
J.Med.Chem., 64, 2021
|
|
7KHH
 
 | | Ternary complex of VHL/BRD4-BD1/Compound9 (4-(3,5-difluoropyridin-2-yl)-N-(11-(((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-11-oxoundecyl)-10-methyl-7-((methylsulfonyl)methyl)-11-oxo-3,4,10,11-tetrahydro-1H-1,4,10-triazadibenzo[cd,f]azulene-6-carboxamide) | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Murray, J.M. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy.
J.Med.Chem., 64, 2021
|
|
8FBX
 
 | | Selenosugar synthase SenB from Variovorax paradoxus | | Descriptor: | CHLORIDE ION, SODIUM ION, Selenosugar synthase SenB | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2022-11-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Characterization and Ligand-Induced Conformational Changes of SenB, a Se-Glycosyltransferase Involved in Selenoneine Biosynthesis.
Biochemistry, 62, 2023
|
|
7PII
 
 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, DNA (122-MER), DNA (123-MER), ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Barford, D. | | Deposit date: | 2021-08-19 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
6MIN
 
 | |
6MIM
 
 | |
6SLG
 
 | | HUMAN ERK2 WITH ERK1/2 INHIBITOR, AZD0364. | | Descriptor: | (6~{R})-7-[[3,4-bis(fluoranyl)phenyl]methyl]-6-(methoxymethyl)-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-5,6-dihydroimidazo[1,2-a]pyrazin-8-one, 1,2-ETHANEDIOL, ERK-tide, ... | | Authors: | Breed, J, Phillips, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a Potent and Selective Oral Inhibitor of ERK1/2 (AZD0364) That Is Efficacious in Both Monotherapy and Combination Therapy in Models of Nonsmall Cell Lung Cancer (NSCLC).
J.Med.Chem., 62, 2019
|
|
6MIO
 
 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9pr | | Descriptor: | Histone H3K9pr, Transcription initiation factor TFIID subunit 14 | | Authors: | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
8FRZ
 
 | |
8FSB
 
 | |
8FSP
 
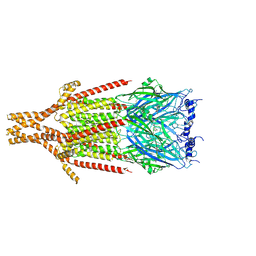 | | Full-length mouse 5-HT3A receptor in complex with SMP100, open-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FSZ
 
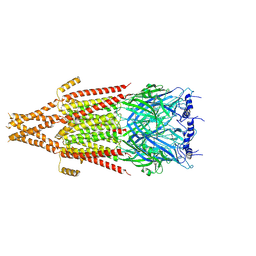 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, open-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FRW
 
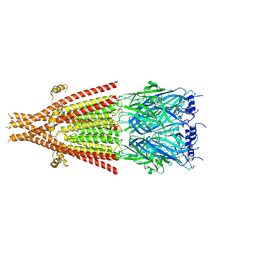 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, pre-activated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-09 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FRX
 
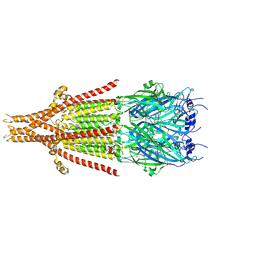 | | Full-length mouse 5-HT3A receptor in complex with SMP100, pre-activated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-09 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6MIQ
 
 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9bu | | Descriptor: | Histone H3K9bu, Transcription initiation factor TFIID subunit 14 | | Authors: | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
8WRH
 
 | | SARS-CoV-2 XBB.1.5.70 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRL
 
 | | XBB.1.5 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
6MIP
 
 | |
6VQF
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244- 487)-L6-SRC1(678-692)) IN COMPLEX WITH AN INVERSE AGONIST | | Descriptor: | (1R,3S,4R)-4-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]-3-methylcyclohexane-1-carboxylic acid, GLYCEROL, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-02-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of BMS-986251: A Clinically Viable, Potent, and Selective ROR gamma t Inverse Agonist.
Acs Med.Chem.Lett., 11, 2020
|
|
8X1V
 
 | |
