8ZGC
 
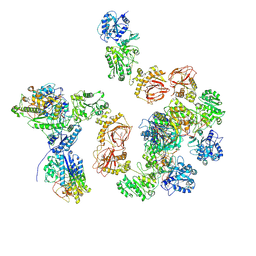 | | Human lysine O-link glycosylation complex, LH3/ColGalT1 with bound UDP-galactose | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Peng, J, Li, W, Yao, D, Xia, Y, Wang, Q, Cai, Y, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Cao, Y. | | Deposit date: | 2024-05-09 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | The structural basis for the human procollagen lysine hydroxylation and dual-glycosylation.
Nat Commun, 16, 2025
|
|
7BX8
 
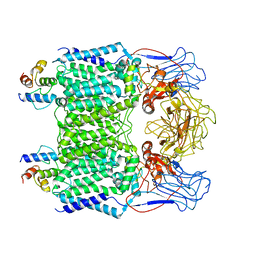 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | Descriptor: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7V4Y
 
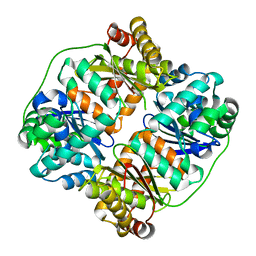 | | TTHA1264/TTHA1265 complex | | Descriptor: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | Authors: | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | Deposit date: | 2021-08-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
7CP9
 
 | | Cryo-EM structure of human mitochondrial translocase TOM complex at 3.0 angstrom. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Guan, Z, Yan, L, Wang, Q, Yan, C, Yin, P. | | Deposit date: | 2020-08-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into assembly of human mitochondrial translocase TOM complex.
Cell Discov, 7, 2021
|
|
7CZB
 
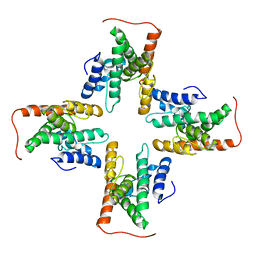 | | The cryo-EM structure of the ERAD retrotranslocation channel formed by human Derlin-1 | | Descriptor: | Derlin-1 | | Authors: | Rao, B, Li, S, Yao, D, Wang, Q, Xia, Y, Jia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2020-09-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The cryo-EM structure of an ERAD protein channel formed by tetrameric human Derlin-1.
Sci Adv, 7, 2021
|
|
7BWR
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | Authors: | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | Deposit date: | 2020-07-01 | | Release date: | 2020-10-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
7VZF
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
7U4K
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with ML162 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(3-chloro-4-methoxyphenyl)-N-[(1R)-2-oxo-2-[(2-phenylethyl)amino]-1-(thiophen-2-yl)ethyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4L
 
 | | Crystal structure of human GPX4-U46C in complex with MAC-5576 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, thiophene-2-carbaldehyde | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4N
 
 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4M
 
 | | Crystal structure of human GPX4-U46C in complex with LOC1886 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole-2-carbaldehyde, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4J
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with TMT10 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4I
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with CDS9 | | Descriptor: | 2-bromo-N-[(thiophen-2-yl)methyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
9LKT
 
 | | Human RNA Polymerase III de novo transcribing complex 10 (TC10) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (123-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q.M, Ren, Y.L, Jin, Q.W, Chen, X.Z, Xu, Y.H. | | Deposit date: | 2025-01-16 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
6LZG
 
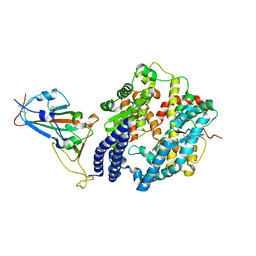 | | Structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Wang, Q.H, Song, H, Qi, J.X. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2.
Cell, 181, 2020
|
|
6O61
 
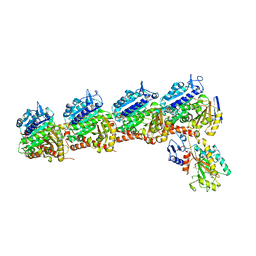 | | Tubulin-RB3_SLD-TTL in complex with compound ABI-231 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structure-Guided Design, Synthesis, and Biological Evaluation of (2-(1H-Indol-3-yl)-1H-imidazol-4-yl)(3,4,5-trimethoxyphenyl) Methanone (ABI-231) Analogues Targeting the Colchicine Binding Site in Tubulin.
J.Med.Chem., 62, 2019
|
|
6LH8
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
6LHZ
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
6D88
 
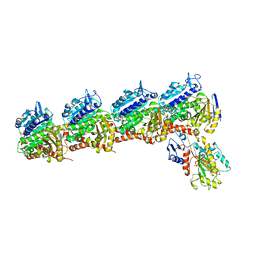 | | Tubulin-RB3_SLD-TTL in complex with compound 13f | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2018-04-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural Modification of the 3,4,5-Trimethoxyphenyl Moiety in the Tubulin Inhibitor VERU-111 Leads to Improved Antiproliferative Activities.
J. Med. Chem., 61, 2018
|
|
6O5M
 
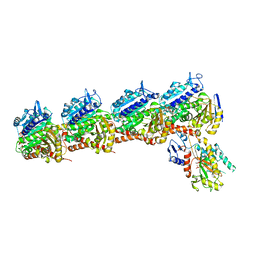 | | Tubulin-RB3_SLD-TTL in complex with compound 10bb | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design, Synthesis, and Biological Evaluation of (2-(1H-Indol-3-yl)-1H-imidazol-4-yl)(3,4,5-trimethoxyphenyl) Methanone (ABI-231) Analogues Targeting the Colchicine Binding Site in Tubulin.
J.Med.Chem., 62, 2019
|
|
6O5N
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 10ab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structure-Guided Design, Synthesis, and Biological Evaluation of (2-(1H-Indol-3-yl)-1H-imidazol-4-yl)(3,4,5-trimethoxyphenyl) Methanone (ABI-231) Analogues Targeting the Colchicine Binding Site in Tubulin.
J.Med.Chem., 62, 2019
|
|
5GZR
 
 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
4XI2
 
 | |
6L69
 
 | | Crystal structure of CYP154C2 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regio- and stereoselective hydroxylation of testosterone by a novel cytochrome P450 154C2 from Streptomyces avermitilis.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
