4Y2C
 
 | | M300V 3D polymerase mutant of EMCV | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
6G76
 
 | |
6G78
 
 | | RSK4 N-terminal Kinase Domain S232E in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-6 | | Authors: | Prischi, F, Ali, M.M. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposed floxacins targeting RSK4 prevent chemoresistance and metastasis in lung and bladder cancer.
Sci Transl Med, 13, 2021
|
|
6G77
 
 | | RSK4 N-terminal Kinase Domain in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-6, ZINC ION | | Authors: | Prischi, F, Ali, M.M. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Repurposed floxacins targeting RSK4 prevent chemoresistance and metastasis in lung and bladder cancer.
Sci Transl Med, 13, 2021
|
|
6XKJ
 
 | | Cryo-EM structure of CARD8-CARD filament | | Descriptor: | Caspase recruitment domain-containing protein 8 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | Deposit date: | 2020-06-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
6XKK
 
 | | Cryo-EM structure of the NLRP1-CARD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | Deposit date: | 2020-06-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
6M3B
 
 | | hAPC-c25k23 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, ... | | Authors: | Wang, X, Li, L, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
6M3C
 
 | | hAPC-h1573 Fab complex | | Descriptor: | Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, h1573 Fab H chain, ... | | Authors: | Wang, X, Wang, D, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
5LGN
 
 | | Thieno[3,2-b]pyrrole-5-carboxamides as Novel Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1: Compound 19 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Mattevi, A, Ciossani, G. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 1: High-Throughput Screening and Preliminary Exploration.
J. Med. Chem., 60, 2017
|
|
5WMD
 
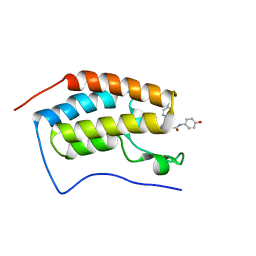 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(4-hydroxyphenyl)acetamide, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5WMG
 
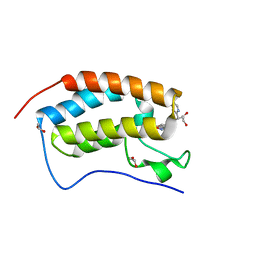 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | Descriptor: | 1,2-ETHANEDIOL, 4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1S)-1-(pyridin-2-yl)ethyl]-1H-pyrrolo[3,2-b]pyridin-3-yl}benzoic acid, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5WMA
 
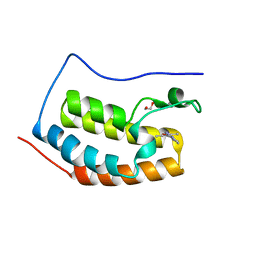 | | N-terminal bromodomain of BRD4 in complex with PLX5981 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
8QFZ
 
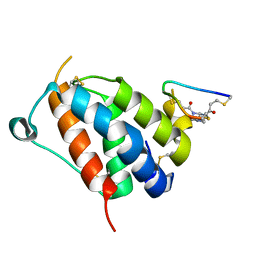 | | TSLP-Bicycle complex | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, CYS-HIS-TRP-LEU-GLU-ASN-CYS-TRP-ARG-GLY-PHE-CYS, Thymic stromal lymphopoietin | | Authors: | Petersen, J. | | Deposit date: | 2023-09-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Characterization of a Bicyclic Peptide (Bicycle) Binder to Thymic Stromal Lymphopoietin.
J.Med.Chem., 67, 2024
|
|
4WFN
 
 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 in complex with erythromycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-16 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
6ZQO
 
 | | EYFP mutant - F165G | | Descriptor: | G protein/GFP fusion protein, SULFATE ION | | Authors: | Pletnev, V.Z, Pletnev, S.V, Pletneva, N.V. | | Deposit date: | 2020-07-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Amino acid residue at the 165th position tunes EYFP chromophore maturation. A structure-based design.
Comput Struct Biotechnol J, 19, 2021
|
|
6RJI
 
 | | X-ray structure of the elongation factor P of S. aureus | | Descriptor: | Elongation factor P | | Authors: | Fatkhullin, B.F, Golubev, A.A, Gabdulkhakov, A.G, Khusainov, I.S, Validov, S.Z, Usachev, K.S, Yusupova, G, Yusupov, M.M. | | Deposit date: | 2019-04-27 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | NMR and crystallographic structural studies of the Elongation factor P from Staphylococcus aureus.
Eur.Biophys.J., 49, 2020
|
|
7A0S
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
7A0R
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L15, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
7A18
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin IV | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L15, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
6RK3
 
 | | Solution structure of the ribosome Elongation Factor P (EF-P) from Staphylococcus aureus | | Descriptor: | Elongation factor P | | Authors: | Usachev, K, Fatkhullin, B, Gabdulkhakov, A, Khusainov, I, Golubev, A, Validov, S, Yusupova, G, Yusupov, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and crystallographic structural studies of the Elongation factor P from Staphylococcus aureus.
Eur.Biophys.J., 49, 2020
|
|
8BYV
 
 | | Cryo-EM structure of a Staphylococus aureus 30S-RbfA complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Bikmullin, A.G, Fatkhullin, B, Stetsenko, A, Guskov, A, Yusupov, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
7B42
 
 | | Crystal structure of c-MET bound by compound 8 | | Descriptor: | 3-[(3-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3Q
 
 | | Crystal structure of c-MET bound by compound 1 | | Descriptor: | 1-(phenylmethyl)-5~{H}-pyrrolo[3,2-c]pyridin-4-one, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B40
 
 | | Crystal structure of c-MET bound by compound 6 | | Descriptor: | 3-(phenylmethyl)-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3W
 
 | | Crystal structure of c-MET bound by compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-fluoranyl-1~{H}-indazol-4-yl)-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
