3SQE
 
 | |
3MVT
 
 | | Crystal structure of apo mADA at 2.2A resolution | | Descriptor: | Adenosine deaminase, CHLORIDE ION, GLYCEROL | | Authors: | Niu, W, Shu, Q, Chen, Z, Mathews, S, Di Cera, E, Frieden, C. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The role of Zn2+ on the structure and stability of murine adenosine deaminase.
J.Phys.Chem.B, 114, 2010
|
|
5X0Y
 
 | | Complex of Snf2-Nucleosome complex with Snf2 bound to SHL2 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
3SJ8
 
 | | Crystal structure of the 3C protease from coxsackievirus A16 | | Descriptor: | 3C protease | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3EDX
 
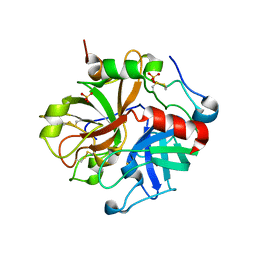 | | Crystal structure of the W215A/E217A mutant of murine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-pelc, L, Di Cera, E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for the Kinetic Differences of the W215A/E217A Mutant of Human and Murine Thrombin
To be Published
|
|
3SJO
 
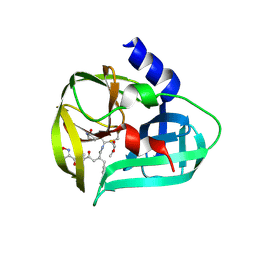 | | structure of EV71 3C in complex with Rupintrivir (AG7088) | | Descriptor: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
4N0N
 
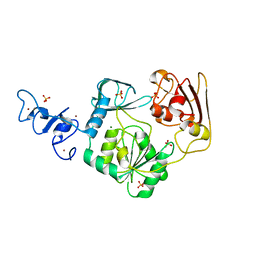 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | Descriptor: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
3S6P
 
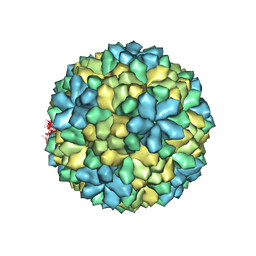 | |
3SJK
 
 | | Crystal structure of the C147A mutant 3C from enterovirus 71 | | Descriptor: | 3C protease, KPVLRTATVQGPSLDF peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
2PUX
 
 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 3, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5EDM
 
 | | Crystal structure of prothrombin deletion mutant residues 154-167 ( Form I ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
2PV9
 
 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 4, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3SQH
 
 | |
5X0X
 
 | | Complex of Snf2-Nucleosome complex with Snf2 bound to position +6 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
4N0O
 
 | | Complex structure of Arterivirus nonstructural protein 10 (helicase) with DNA | | Descriptor: | CALCIUM ION, DNA, Replicase polyprotein 1ab, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
5Z3L
 
 | | Structure of Snf2-nucleosome complex in apo state | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5YYL
 
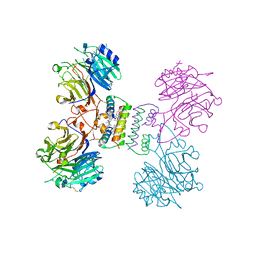 | | Structure of Major Royal Jelly Protein 1 Oligomer | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tian, W, Chen, Z. | | Deposit date: | 2017-12-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
5Z3V
 
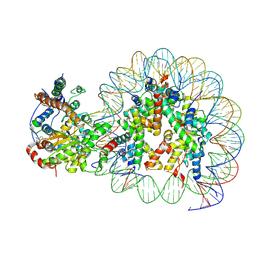 | | Structure of Snf2-nucleosome complex at shl-2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3O
 
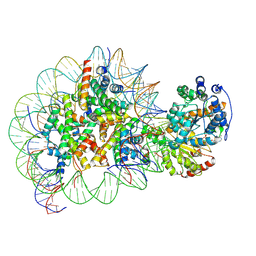 | | Structure of Snf2-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z38
 
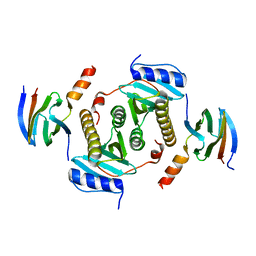 | | Crystal structure of CsrA bound to CesT | | Descriptor: | CesT protein, Truncated-CsrA, wild type CsrA | | Authors: | Ye, F, Yang, F, Yu, R, Lin, X, Qi, J, Chen, Z, Gao, G.F, Lu, G. | | Deposit date: | 2018-01-05 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT
Nat Commun, 9, 2018
|
|
5Z3U
 
 | | Structure of Snf2-nucleosome complex at shl2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
3MVI
 
 | | Crystal structure of holo mADA at 1.6 A resolution | | Descriptor: | Adenosine deaminase, GLYCEROL, ZINC ION | | Authors: | Niu, W, Shu, Q, Chen, Z, Mathews, S, Di Cera, E, Frieden, C. | | Deposit date: | 2010-05-04 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The role of Zn2+ on the structure and stability of murine adenosine deaminase.
J.Phys.Chem.B, 114, 2010
|
|
4NZQ
 
 | | Crystal structure of Ca2+-free prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O67
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with GAMP | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O68
 
 | | Structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
