1B9L
 
 | | 7,8-DIHYDRONEOPTERIN TRIPHOSPHATE EPIMERASE | | Descriptor: | PROTEIN (EPIMERASE) | | Authors: | Ploom, T, Haussmann, C, Hof, P, Steinbacher, S, Bacher, A, Richardson, J, Huber, R. | | Deposit date: | 1999-02-11 | | Release date: | 2000-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 7,8-dihydroneopterin triphosphate epimerase.
Structure Fold.Des., 7, 1999
|
|
1ONN
 
 | | IspC apo structure | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
1OXD
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1OXE
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1CFR
 
 | |
1CPC
 
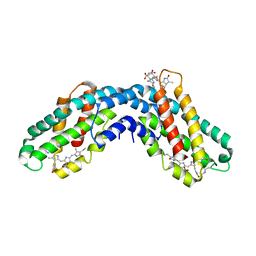 | | ISOLATION, CRYSTALLIZATION, CRYSTAL STRUCTURE ANALYSIS AND REFINEMENT OF CONSTITUTIVE C-PHYCOCYANIN FROM THE CHROMATICALLY ADAPTING CYANOBACTERIUM FREMYELLA DIPLOSIPHON AT 1.66 ANGSTROMS RESOLUTION | | Descriptor: | C-PHYCOCYANIN (ALPHA SUBUNIT), C-PHYCOCYANIN (BETA SUBUNIT), PHYCOCYANOBILIN | | Authors: | Duerring, M, Schmidt, G.B, Huber, R. | | Deposit date: | 1990-10-11 | | Release date: | 1993-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Isolation, crystallization, crystal structure analysis and refinement of constitutive C-phycocyanin from the chromatically adapting cyanobacterium Fremyella diplosiphon at 1.66 A resolution.
J.Mol.Biol., 217, 1991
|
|
1ORV
 
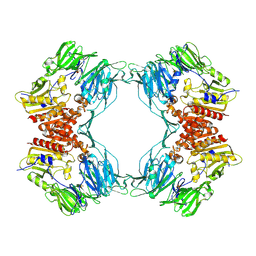 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1FFU
 
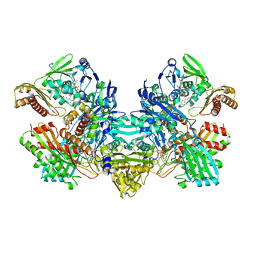 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA WHICH LACKS THE MO-PYRANOPTERIN MOIETY OF THE MOLYBDENUM COFACTOR | | Descriptor: | CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, CUTM, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
1PCA
 
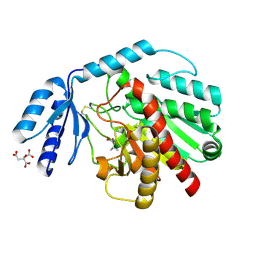 | | THREE DIMENSIONAL STRUCTURE OF PORCINE PANCREATIC PROCARBOXYPEPTIDASE A. A COMPARISON OF THE A AND B ZYMOGENS AND THEIR DETERMINANTS FOR INHIBITION AND ACTIVATION | | Descriptor: | CITRIC ACID, PROCARBOXYPEPTIDASE A PCPA, VALINE, ... | | Authors: | Guasch, A, Coll, M, Aviles, F.X, Huber, R. | | Deposit date: | 1991-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of porcine pancreatic procarboxypeptidase A. A comparison of the A and B zymogens and their determinants for inhibition and activation.
J.Mol.Biol., 224, 1992
|
|
1G0U
 
 | | A GATED CHANNEL INTO THE PROTEASOME CORE PARTICLE | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Bajorek, M, Kohler, A, Moroder, L, Rubin, D.M, Huber, R, Glickman, M.H, Finley, D. | | Deposit date: | 2000-10-09 | | Release date: | 2000-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A gated channel into the proteasome core particle.
Nat.Struct.Biol., 7, 2000
|
|
1ICQ
 
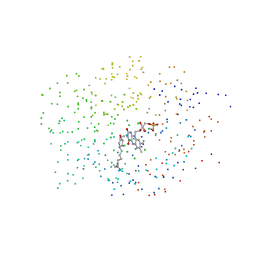 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1I48
 
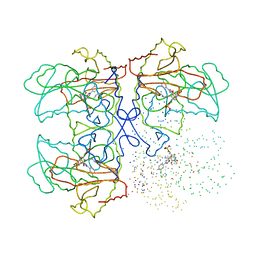 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR CTCPO | | Descriptor: | CARBOXYMETHYLTHIO-3-(3-CHLOROPHENYL)-1,2,4-OXADIAZOL, CYSTATHIONINE GAMMA-SYNTHASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-20 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1FB1
 
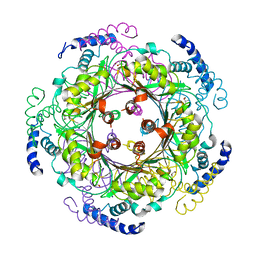 | | CRYSTAL STRUCTURE OF HUMAN GTP CYCLOHYDROLASE I | | Descriptor: | GTP CYCLOHYDROLASE I, ISOPROPYL ALCOHOL, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, G, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-14 | | Release date: | 2000-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FBX
 
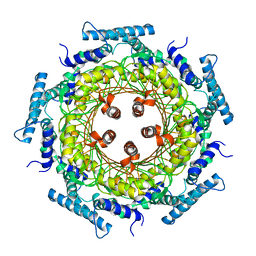 | | CRYSTAL STRUCTURE OF ZINC-CONTAINING E.COLI GTP CYCLOHYDROLASE I | | Descriptor: | CHLORIDE ION, GTP CYCLOHYDROLASE I, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, A, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-17 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1J2Q
 
 | | 20S proteasome in complex with calpain-Inhibitor I from archaeoglobus fulgidus | | Descriptor: | 2-ACETYLAMINO-4-METHYL-PENTANOIC ACID [1-(1-FORMYL-PENTYLCARBAMOYL)-3-METHYL-BUTYL]-AMIDE, Proteasome alpha subunit, Proteasome beta subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
1ICP
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH PEG400 | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1Q62
 
 | | PKA double mutant model of PKB | | Descriptor: | cAMP-dependent protein kinase inhibitor, alpha form, cAMP-dependent protein kinase, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-09-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1ICS
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1I41
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR APPA | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-IMINO]-5-PHOSPHONO-PENT-3-ENOIC ACID, CYSTATHIONINE GAMMA-SYNTHASE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1I43
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR PPCA | | Descriptor: | 3-(PHOSPHONOMETHYL)PYRIDINE-2-CARBOXYLIC ACID, CYSTATHIONINE GAMMA-SYNTHASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1Q24
 
 | | PKA double mutant model of PKB in complex with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent Protein Kinase Inhibitor, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-07-23 | | Release date: | 2003-08-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1G8H
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE TERNARY PRODUCT COMPLEX WITH APS AND PPI | | Descriptor: | ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1J2P
 
 | | alpha-ring from the proteasome from archaeoglobus fulgidus | | Descriptor: | Proteasome alpha subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
